Simultaneous Down-Regulation of Dominant Cinnamoyl CoA Reductase and Cinnamyl Alcohol Dehydrogenase Dramatically Altered Lignin Content in Mulberry
- PMID: 39771210
- PMCID: PMC11676671
- DOI: 10.3390/plants13243512
Simultaneous Down-Regulation of Dominant Cinnamoyl CoA Reductase and Cinnamyl Alcohol Dehydrogenase Dramatically Altered Lignin Content in Mulberry
Abstract
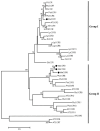
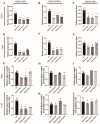
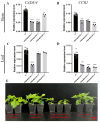
Mulberry (Morus alba L.) is a significant economic tree species in China. The lignin component serves as a critical limiting factor that impacts both the forage quality and the conversion efficiency of mulberry biomass into biofuel. Cinnamoyl CoA reductase (CCR; EC 1.21.1.44) and cinnamyl alcohol dehydrogenase (CAD; EC 1.1.1.95) are the key enzymes that catalyze the final two reductive steps in the biosynthesis of monolignols. In this study, we conducted a comprehensive functional analysis to validate the predominant CCR genes involved in monolignol biosynthesis. In this study, we initially validated the predominant CCR genes implicated in monolignol biosynthesis through an extensive functional analysis. Phylogenetic analysis, tissue-specific expression profiling and enzymatic assays indicated that MaCCR1 is the authentic CCR involved in lignin biosynthesis. Furthermore, the expression level of MaCCR1 exhibited a significant positive correlation with lignin content, and the down-regulation of MaCCR1 via virus-induced gene silencing resulted in altered lignin content in mulberry. The down-regulation of MaCCR1 and MaCAD3/4, both individually and concurrently, exhibited markedly different effects on lignin content and mulberry growth. Specifically, the simultaneous down-regulation of MaCCR1 and MaCAD3/4 significantly altered lignin content in mulberry, resulting in dwarfism of the plants. Conversely, the down-regulation of MaCAD3/4 alone not only decreased lignin content but also led to an increase in biomass. These findings offer compelling evidence elucidating the roles of MaCCRs in mulberry and identify specific target genes, thereby providing a crucial foundation for the genetic modification of lignin biosynthesis.
Keywords: Morus; VIGS; cinnamoyl CoA reductase; cinnamyl alcohol dehydrogenase; enzymatic assay; lignin.
Conflict of interest statement
The authors declare that the research was conducted in absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Systematic functional characterization of cinnamyl alcohol dehydrogenase family members revealed their functional divergence in lignin biosynthesis and stress responses in mulberry.Plant Physiol Biochem. 2022 Sep 1;186:145-156. doi: 10.1016/j.plaphy.2022.07.008. Epub 2022 Jul 11. Plant Physiol Biochem. 2022. PMID: 35849944
-
Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity.Phytochemistry. 2002 Oct;61(3):221-94. doi: 10.1016/s0031-9422(02)00211-x. Phytochemistry. 2002. PMID: 12359514 Review.
-
The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana.Mol Plant. 2011 Jan;4(1):70-82. doi: 10.1093/mp/ssq045. Epub 2010 Sep 9. Mol Plant. 2011. PMID: 20829305
-
Clade classification of monolignol biosynthesis gene family members reveals target genes to decrease lignin in Lolium perenne.Plant Biol (Stuttg). 2015 Jul;17(4):877-92. doi: 10.1111/plb.12316. Epub 2015 Mar 13. Plant Biol (Stuttg). 2015. PMID: 25683375
-
The polymorphism of the genes/enzymes involved in the last two reductive steps of monolignol synthesis: what is the functional significance?C R Biol. 2004 Sep-Oct;327(9-10):837-45. doi: 10.1016/j.crvi.2004.04.007. C R Biol. 2004. PMID: 15587075 Review.
References
-
- Łochyńska M. Energy and nutritional properties of the white mulberry (Morus alba L.) J. Agric. Sci. Technol. 2015;5:709–716.
Grants and funding
- BK20210879/Natural Science Foundation of Jiangsu Province
- NSF 32201526/National Natural Science Foundation
- 21KJB180013/Natural Science Research of Jiangsu Higher Education Institutions of China
- 19200382/Crop Germplasm Resources Protection Project of the Ministry of Agriculture and Rural Affairs of the People's Republic of China
- NCGRC-2020-041/National Infrastructure for Crop Germplasm Resources
LinkOut - more resources
Full Text Sources
Miscellaneous

