Unraveling the Molecular Mechanisms of Blueberry Root Drought Tolerance Through Yeast Functional Screening and Metabolomic Profiling
- PMID: 39771226
- PMCID: PMC11678528
- DOI: 10.3390/plants13243528
Unraveling the Molecular Mechanisms of Blueberry Root Drought Tolerance Through Yeast Functional Screening and Metabolomic Profiling
Abstract
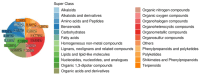
Blueberry plants are among the most important fruit-bearing shrubs, but they have shallow, hairless roots that are not conducive to water and nutrient uptake, especially under drought conditions. Therefore, the mechanism underlying blueberry root drought tolerance should be clarified. Hence, we established a yeast expression library comprising blueberry genes associated with root responses to drought stress. High-throughput sequencing technology enabled the identification of 1475 genes potentially related to drought tolerance. A subsequent KEGG enrichment analysis revealed 77 key genes associated with six pathways: carbon and energy metabolism, biosynthesis of secondary metabolites, nucleotide and amino acid metabolism, genetic information processing, signal transduction, and material transport and catabolism. Metabolomic profiling of drought-tolerant yeast strains under drought conditions detected 1749 differentially abundant metabolites (DAMs), including several up-regulated metabolites (organic acids, amino acids and derivatives, alkaloids, and phenylpropanoids). An integrative analysis indicated that genes encoding several enzymes, including GALM, PK, PGLS, and PIP5K, modulate key carbon metabolism-related metabolites, including D-glucose 6-phosphate and β-D-fructose 6-phosphate. Additionally, genes encoding FDPS and CCR were implicated in terpenoid and phenylalanine biosynthesis, which affected metabolite contents (e.g., farnesylcysteine and tyrosine). Furthermore, genes for GST and GLT1, along with eight DAMs, including L-γ-glutamylcysteine and L-ornithine, contributed to amino acid metabolism, while genes encoding NDPK and APRT were linked to purine metabolism, thereby affecting certain metabolites (e.g., inosine and 3',5'-cyclic GMP). Overall, the yeast functional screening system used in this study effectively identified genes and metabolites influencing blueberry root drought tolerance, offering new insights into the associated molecular mechanisms.
Keywords: blueberry root; drought tolerance; genes; metabolomic profiling; molecular mechanisms; yeast functional screening.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures










Similar articles
-
Comparative metabolomics of root-tips reveals distinct metabolic pathways conferring drought tolerance in contrasting genotypes of rice.BMC Genomics. 2023 Mar 27;24(1):152. doi: 10.1186/s12864-023-09246-z. BMC Genomics. 2023. PMID: 36973662 Free PMC article.
-
Transcriptomic and Metabolomic Profiling of Root Tissue in Drought-Tolerant and Drought-Susceptible Wheat Genotypes in Response to Water Stress.Int J Mol Sci. 2024 Sep 27;25(19):10430. doi: 10.3390/ijms251910430. Int J Mol Sci. 2024. PMID: 39408761 Free PMC article.
-
Molecular mechanisms regulating glucose metabolism in quinoa (Chenopodium quinoa Willd.) seeds under drought stress.BMC Plant Biol. 2024 Aug 23;24(1):796. doi: 10.1186/s12870-024-05510-w. BMC Plant Biol. 2024. PMID: 39174961 Free PMC article.
-
Metabolomic study reveals key metabolic adjustments in the xerohalophyte Salvadora persica L. during adaptation to water deficit and subsequent recovery conditions.Plant Physiol Biochem. 2020 May;150:180-195. doi: 10.1016/j.plaphy.2020.02.036. Epub 2020 Feb 28. Plant Physiol Biochem. 2020. PMID: 32146282
-
Review on blueberry drought tolerance from the perspective of cultivar improvement.Front Plant Sci. 2024 May 14;15:1352768. doi: 10.3389/fpls.2024.1352768. eCollection 2024. Front Plant Sci. 2024. PMID: 38807786 Free PMC article. Review.
Cited by
-
Abiotic and Biotic Stress of the Crops and Horticultural Plants.Plants (Basel). 2025 Jun 21;14(13):1910. doi: 10.3390/plants14131910. Plants (Basel). 2025. PMID: 40647919 Free PMC article.
References
-
- Yasmeen T., Arif M.S., Tariq M., Akhtar S., Syrish A., Haidar W., Rizwan M., Hussain M.I., Ahmad A., Ali S. Biofilm producing plant growth promoting bacteria in combination with glycine betaine uplift drought stress tolerance of maize plant. Front. Plant Sci. 2024;15:1327552. doi: 10.3389/fpls.2024.1327552. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

