Synthesis and Degradation of the Phytohormone Indole-3-Acetic Acid by the Versatile Bacterium Paraburkholderia xenovorans LB400 and Its Growth Promotion of Nicotiana tabacum Plant
- PMID: 39771231
- PMCID: PMC11676955
- DOI: 10.3390/plants13243533
Synthesis and Degradation of the Phytohormone Indole-3-Acetic Acid by the Versatile Bacterium Paraburkholderia xenovorans LB400 and Its Growth Promotion of Nicotiana tabacum Plant
Abstract
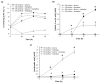
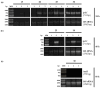
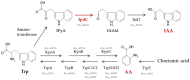
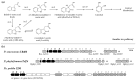
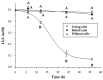
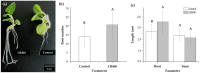
Plant growth-promoting bacteria (PGPB) play a role in stimulating plant growth through mechanisms such as the synthesis of the phytohormone indole-3-acetic acid (IAA). The aims of this study were the characterization of IAA synthesis and degradation by the model aromatic-degrading bacterium Paraburkholderia xenovorans LB400, and its growth promotion of the Nicotiana tabacum plant. Strain LB400 was able to synthesize IAA (measured by HPLC) during growth in the presence of tryptophan and at least one additional carbon source; synthesis of anthranilic acid was also observed. RT-PCR analysis indicates that under these conditions, strain LB400 expressed the ipdC gene, which encodes indole-3-pyruvate decarboxylase, suggesting that IAA biosynthesis proceeds through the indole-3-pyruvate pathway. In addition, strain LB400 degraded IAA and grew on IAA as a sole carbon and energy source. Strain LB400 expressed the iacC and catA genes, which encode the α subunit of the aromatic-ring-hydroxylating dioxygenase in the IAA catabolic pathway and the catechol 1,2-dioxygenase, respectively, which may suggest a peripheral IAA pathway leading to the central catechol pathway. Notably, P. xenovorans LB400 promoted the growth of tobacco seedlings, increasing the number and the length of the roots. In conclusion, this study indicates that the versatile bacterium P. xenovorans LB400 is a PGPB.
Keywords: IAA degradation; IAA synthesis; Paraburkholderia xenovorans LB400; indole-3-acetic acid (IAA); plant growth-promoting bacteria (PGPB).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Vega-Celedón P., Bravo G., Velásquez A., Cid F.P., Valenzuela M., Ramírez I., Vasconez I.N., Álvarez I., Jorquera M.A., Seeger M. Microbial diversity of psychrotolerant bacteria isolated from wild flora of Andes Mountains and Patagonia of Chile towards the selection of plant growth-promoting bacterial consortia to alleviate cold stress in plants. Microorganisms. 2021;9:538. doi: 10.3390/microorganisms9030538. - DOI - PMC - PubMed
-
- Khoso M., Wagan S., Alam I., Hussain A., Qurban A., Sudipta S., Poudel T., Manghwar H., Liu F. Impact of plant growth-promoting rhizobacteria (PGPR) on plant nutrition and root characteristics: Current perspective. Plant Stress. 2024;11:10034. doi: 10.1016/j.stress.2023.100341. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

