Comparative Physiological and Transcriptomics Profiling Provides Integrated Insight into Melatonin Mediated Salt and Copper Stress Tolerance in Selenicereus undatus L
- PMID: 39771301
- PMCID: PMC11678089
- DOI: 10.3390/plants13243602
Comparative Physiological and Transcriptomics Profiling Provides Integrated Insight into Melatonin Mediated Salt and Copper Stress Tolerance in Selenicereus undatus L
Abstract
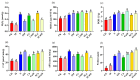
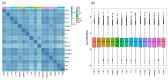
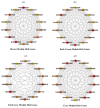
Selenicereus undatus L., (pitaya) is an important tropical fruit crop, and faces significant challenges from soil salinity and heavy metal toxicity. This study explores the role of melatonin (M) in enhancing stress tolerance in pitaya against salinity (S) and copper (Cu) toxicity, both individually and in combination (SCu). SCu stress reduced plant biomass by ~54%, while melatonin application mitigated stress effects and increased plant growth by ~73.26% under SCuM compared to SCu treatment. Antioxidant activities were also modulated by stress. Transcriptomic analysis revealed 21 differentially expressed genes (DEGs) common across stress treatments and 13 DEGs specific to combined melatonin with stress treatments involved in stress signaling, secondary metabolite biosynthesis, and photosynthesis. A weighted gene co-expression network analysis (WGCNA) identified four gene modules (brown, dark green, dark grey, and grey) significantly associated with phenotypic traits. A protein-protein interaction (PPI) network analysis highlighted 14 hub genes per module, including GH3, JAZ, PAL, CCR, and POD, implicated in MAPK signaling, phenylpropanoid biosynthesis, and hormone signaling pathways. Integration of DESeq2 and WGCNA identified 12 key stress-responsive genes strongly correlated with phenotypic traits. This study provides insights into regulatory mechanisms underlying stress responses and highlights candidate genes for developing stress-resilient S. undatus through breeding programs.
Keywords: Selenicereus undatus L.; WGCNA; abiotic stresses; physiology; transcriptomic.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








References
-
- Chen J., Xie F., Cui Y., Chen C., Lu W., Hu X., Hua Q., Zhao J., Wu Z., Gao D., et al. A chromosome-scale genome sequence of pitaya (Hylocereus undatus) provides novel insights into the genome evolution and regulation of betalain biosynthesis. Hortic. Res. 2021;8:164. doi: 10.1038/s41438-021-00612-0. - DOI - PMC - PubMed
-
- Nerd A., Tel-Zur N., Mizrahi Y. Fruits of vine and columnar cacti. In: Nobel P.S., editor. Cacti, Biology and Uses. University of California Press; Los Angeles, CA, USA: 2002. pp. 185–198.
-
- Khan D., Harris A., Zaman Q.U., Wang H.X., Wen J., Landis J.B., Wang H.F. The evolutionary history and distribution of cactus germplasm resources, as well as potential domestication under a changing climate. J. Syst. Evol. 2024;62:858–875. doi: 10.1111/jse.13042. - DOI
-
- Le Bellec F., Vaillant F., Imbert E. Pitahaya (Hylocereus spp.): A new fruit crop, a market with a future. Fruits. 2006;61:237–250. doi: 10.1051/fruits:2006021. - DOI
-
- Ibrahim S.R.M., Mohamed G.A., Khedr A.I.M., Zayed M.F., El-Kholy A.A.E.S. Genus Hylocereus: Beneficial phytochemicals, nutritional importance, and biological relevance—A review. J. Food Biochem. 2018;42:e12491. doi: 10.1111/jfbc.12491. - DOI
Grants and funding
- ZDYF2022XDNY190/Hainan Province Science and Technology Special Fund
- SCKJ-JYRC-2022-83/Project of Sanya Yazhou Bay Science and Technology City
- 421RC486/Hainan Provincial Natural Science Foundation of China
- XTCX2022NYB09/Collaborative Innovation Center of Nanfan and High Efficiency Tropical Agriculture
LinkOut - more resources
Full Text Sources

