Affinity Peptide-Based Circularly Permuted Fluorescent Protein Biosensors for Non-Small Cell Lung Cancer Diagnosis
- PMID: 39771637
- PMCID: PMC11679068
- DOI: 10.3390/s24247899
Affinity Peptide-Based Circularly Permuted Fluorescent Protein Biosensors for Non-Small Cell Lung Cancer Diagnosis
Abstract
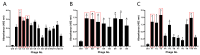
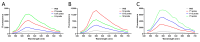
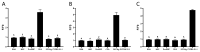
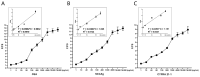
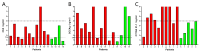
Non-small cell lung cancer (NSCLC) is the predominant form of lung cancer and poses a significant public health challenge. Early detection is crucial for improving patient outcomes, with serum biomarkers such as carcinoembryonic antigen (CEA), squamous cell carcinoma antigen (SCCAg), and cytokeratin fragment 19 (CYFRA 21-1) playing a critical role in early screening and pathological classification of NSCLC. However, due to being mainly based on corresponding antibody binding reactions, existing detection technologies for these serum biomarkers have shortcomings such as complex operations, high false positive rates, and high costs. This study aimed to develop new methods for detecting CEA, SCCAg, and CYFRA 21-1 to assist in the diagnosis of NSCLC. Affinity peptides of CEA, SCCAg, and CYFRA 21-1, respectively, were screened by phage display technology, and the peptides' binding affinities were determined by enzyme-linked immunosorbent assay and biolayer interferometry. Peptides with high affinity were then integrated as binding domains into biosensors by fusing them with circularly permuted fluorescent proteins (cpFPs) through genetic coding. The resulting biosensors, C4 biosensor for CEA, S1 biosensor for SCCAg, and Y3 biosensor for CYFRA 21-1, demonstrated robust sensitivity and specificity even at concentrations as low as 1 ng/mL for their respective tumor markers. When applied to clinical samples and recalibrated for the upper limit of normal concentrations, the biosensors exhibited enhanced sensitivity and specificity for NSCLC diagnosis. This study introduced innovative biosensors for the detection of CEA, SCCAg, and CYFRA 21-1, providing a highly sensitive, specific, rapid, and cost-effective diagnostic alternative that could significantly improve NSCLC screening rates.
Keywords: carcinoembryonic antigen; circularly permuted fluorescent protein; cytokeratin fragment 19; non-small cell lung cancer; phage display; squamous cell carcinoma antigen.
Conflict of interest statement
Author B.W. was employed by China Certification and Inspection Group Liaoning Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
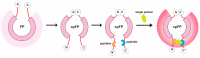


References
-
- China NHCotPsRo Guidelines for the Diagnosis and Treatment of Primary Lung Cancer. Chin. J. Ration. Drug Use. 2022;9:1–28.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

