Protective Antimicrobial Effect of the Potential Vaccine Created on the Basis of the Structure of the IgA1 Protease from Neisseria meningitidis
- PMID: 39772017
- PMCID: PMC11680179
- DOI: 10.3390/vaccines12121355
Protective Antimicrobial Effect of the Potential Vaccine Created on the Basis of the Structure of the IgA1 Protease from Neisseria meningitidis
Abstract
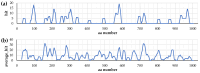
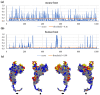
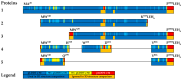
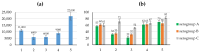
Background/Objectives: IgA1 protease is one of the virulence factors of Neisseria meningitidis, Haemophilus influenzae and other pathogens causing bacterial meningitis. The aim of this research is to create recombinant proteins based on fragments of the mature IgA1 protease A28-P1004 from N. meningitidis serogroup B strain H44/76. These proteins are potential components of an antimeningococcal vaccine for protection against infections caused by pathogenic strains of N. meningitidis and other bacteria producing serine-type IgA1 proteases. Methods: To obtain promising antigens for creating a vaccine, we designed and obtained several recombinant proteins. These proteins consisted of single or directly connected fragments selected from various regions of the IgA1 protease A28-P1004. The choice of these fragments was based on our calculated data on the distribution of linear and conformational B-cell epitopes and MHC-II T-cell epitopes in the structure of IgA1 protease, taking into account the physicochemical properties of potential compounds and the results of a comparative analysis of the spatial structures of the original IgA1 protease and potential recombinant proteins. We studied the immunogenic and protective effects of the obtained proteins on the BALB/c mice against meningococci of serogroups A, B and C. Results: Proteins MA28-P1004-LEH6, MW140-K833-LEH6, MW329-P1004-LEH6, M(W140-H328)-(W412-D604)-(Y866-P1004)-LEH6 and M(W140-Q299)-(Y866-P1004)-LEH6 have shown the following antibody titers, 103/titer: 11 ± 1, 6 ± 2, 6 ± 1, 9 ± 1 and 22 ± 3, respectively. Also, the last two proteins have shown the best average degree of protection from N. meningitidis serogroups A, B and C, %: 62 ± 6, 63 ± 5, 67 ± 4 respectively for M(W140-H328)-(W412-D604)-(Y866-P1004)-LEH6 and 70 ± 5, 66 ± 6, 83 ± 3 respectively for M(W140-Q299)-(Y866-P1004)-LEH6. Conclusions: We selected two recombinant proteins consisting of two (M(W140-Q299)-(Y866-P1004)-LEH6) or three (M(W140-H328)-(W412-D604)-(Y866-P1004)-LEH6) linked fragments of IgA1 protease A28-P1004 as candidate active component for an antimeningococcal vaccine.
Keywords: IgA1 protease; Neisseria meningitidis; epitopes; fused proteins; meningococcal vaccine; protein structure prediction.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Highly Similar Sequences of Mature IgA1 Proteases from Neisseria meningitidis, Neisseria gonorrhoeae and Haemophilus influenzae.Pathogens. 2022 Jun 28;11(7):734. doi: 10.3390/pathogens11070734. Pathogens. 2022. PMID: 35889980 Free PMC article.
-
Induction of Susceptibility to Disseminated Infection with IgA1 Protease-Producing Encapsulated Pathogens Streptococcus pneumoniae, Haemophilus influenzae Type b, and Neisseria meningitidis.mBio. 2022 Jun 28;13(3):e0055022. doi: 10.1128/mbio.00550-22. Epub 2022 Apr 14. mBio. 2022. PMID: 35420467 Free PMC article.
-
Serological Analysis of Immunogenic Properties of Recombinant Meningococcus IgA1 Protease-Based Proteins.Bull Exp Biol Med. 2016 Jul;161(3):391-4. doi: 10.1007/s10517-016-3422-2. Epub 2016 Aug 6. Bull Exp Biol Med. 2016. PMID: 27496029
-
Characteristics of a new meningococcal serogroup B vaccine, bivalent rLP2086 (MenB-FHbp; Trumenba®).Postgrad Med. 2016 Aug;128(6):548-56. doi: 10.1080/00325481.2016.1203238. Epub 2016 Jul 7. Postgrad Med. 2016. PMID: 27467048 Review.
-
Use of expanded Neisseria meningitidis serogroup B panels with the serum bactericidal antibody assay for the evaluation of meningococcal B vaccine effectiveness.Expert Rev Vaccines. 2023 Jan-Dec;22(1):738-748. doi: 10.1080/14760584.2023.2244596. Expert Rev Vaccines. 2023. PMID: 37622470 Review.
References
-
- Zhigis L.S., Kotelnikova O.V., Zinchenko A.A., Karlinsky D.M., Prokopenko Y.A., Rumsh L.D. IgA1 Protease as a Vaccine Basis for Prevention of Bacterial Meningitis. Russ. J. Bioorg. Chem. 2021;47:805–814. doi: 10.1134/S106816202104021X. - DOI
-
- Bolgiano B., Moran E., Beresford N.J., Gao F., Care R., Desai T., Nordgren I.K., Rudd T.R., Feavers I.M., Bore P., et al. Evaluation of Critical Quality Attributes of a Pentavalent (A, C, Y, W, X) Meningococcal Conjugate Vaccine for Global Use. Pathogens. 2021;10:928. doi: 10.3390/pathogens10080928. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

