Expression of an Efficient Selection Marker Out of a Duplicated Site in the ITRs of a Modified Vaccinia Virus Ankara (MVA)
- PMID: 39772039
- PMCID: PMC11680203
- DOI: 10.3390/vaccines12121377
Expression of an Efficient Selection Marker Out of a Duplicated Site in the ITRs of a Modified Vaccinia Virus Ankara (MVA)
Abstract
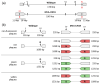
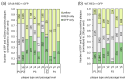
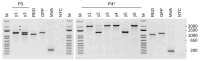
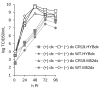
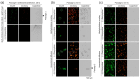
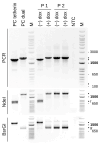
Background/Objectives: Poxviruses are large DNA viruses that replicate in the host cytoplasm without a nuclear phase. As vaccine vectors, they can package and express large recombinant cassettes from different positions of their genomic core region. We present a comparison between wildtype modified vaccinia Ankara (MVA) and isolate CR19, which has significantly expanded inverted terminal repeats (ITRs). With this expansion, a site in wildtype MVA, called deletion site (DS) IV, has been duplicated at both ends of the genome and now occupies an almost central position in the newly formed ITRs. Methods: We inserted various reporter genes into this site and found that the ITRs can be used for transgene expression. However, ITRs are genomic structures that can rapidly adapt to selective pressure through transient duplication and contraction. To test the potential utility of insertions into viral telomers, we inserted a factor from the cellular innate immune system that interferes with viral replication as an example of a difficult transgene. Results: A site almost in the centre of the ITRs can be used for transgene expression, and both sides are mirrored into identical copies. The example of a challenging transgene, tetherin, proved to be surprisingly efficient in selecting candidate vectors against the large background of parental viruses. Conclusions: Insertion of transgenes into ITRs automatically doubles the gene doses. The functionalisation of viruses with tetherin may accelerate the identification and generation of recombinant vectors for personalised medicine and pandemic preparedness.
Keywords: MVA; innate immune system; inverted terminal repeat; tetherin; vaccinia.
Conflict of interest statement
S.A., N.S., A.K., V.S., and I.J. are inventors of patents that are based on the data described in this publication (patent numbers not available at the time of submission of the manuscript). All Authors were employed by the company ProBioGen AG.
Figures


References
-
- Mayr A., Hochstein-Mintzel V., Stickl H. Abstammung, Eigenschaften Und Verwendung Des Attenuierten Vaccinia-Stammes MVA. Infection. 1975;3:6–14. doi: 10.1007/BF01641272. - DOI
-
- Meisinger-Henschel C., Schmidt M., Lukassen S., Linke B., Krause L., Konietzny S., Goesmann A., Howley P., Chaplin P., Suter M., et al. Genomic Sequence of Chorioallantois Vaccinia Virus Ankara, the Ancestor of Modified Vaccinia Virus Ankara. J. Gen. Virol. 2007;88:3249–3259. doi: 10.1099/vir.0.83156-0. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

