Pichia pastoris-Derived β-Glucan Capsules as a Delivery System for DNA Vaccines
- PMID: 39772088
- PMCID: PMC11728682
- DOI: 10.3390/vaccines12121428
Pichia pastoris-Derived β-Glucan Capsules as a Delivery System for DNA Vaccines
Abstract
Background/objectives: DNA vaccines are rapidly produced and adaptable to different pathogens, but they face considerable challenges regarding stability and delivery to the cellular target. Thus, effective delivery methods are essential for the success of these vaccines. Here, we evaluated the efficacy of capsules derived from the cell wall of the yeast Pichia pastoris as a delivery system for DNA vaccines.
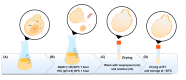
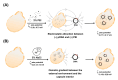
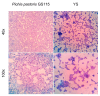
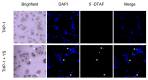
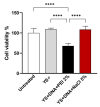
Methods: The capsules were extracted from the yeast Pichia pastoris strain GS115, previously grown in a YPD medium. pVAX1 expression vector was adopted to evaluate the DNA vaccine insertion and delivery. Three encapsulation protocols were tested to identify the most effective in internalizing the plasmid. The presence of plasmids inside the capsules was confirmed by fluorescence microscopy, and the encapsulation efficiency was calculated by the difference between the initial concentration of DNA used for insertion and the concentration of unencapsulated DNA contained in the supernatant. The capsules were subjected to different temperatures to evaluate their thermostability and were co-cultured with macrophages for phagocytosis analysis. HEK-293T cells were adopted to assess the cytotoxicity levels by MTT assay.
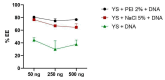
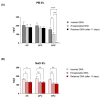
Results: The microscopy results indicated that the macrophages successfully phagocytosed the capsules. Among the protocols tested for encapsulation, the one with 2% polyethylenimine for internalization showed the highest efficiency, with an encapsulation rate above 80%. However, the vaccine capsules obtained with the protocol that used 5% NaCl showed better thermal stability and encapsulation efficiency above 63% without induction of cell viability loss in HEK 293T.
Conclusions: We successfully described a vaccine delivery system using yeast capsules derived from Pichia pastoris, demonstrating its potential for DNA vaccine delivery for the first time. Additional studies will be needed to characterize and improve this delivery strategy.
Keywords: glucan particle; nucleic acids; yeast cell wall.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
Grants and funding
- N° 31/2022, grant number 440315/2022-0/Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq
- APQ-1482-2.02/22/Programa de Apoio à Fixação de Jovens Doutores em Pernambuco
- IBPG-0954-2.00/22/Fundação de Amparo à Ciência e Tecnologia de Pernambuco (FACEPE)
- IBPG-1652-2.00/21/Fundação de Amparo à Ciência e Tecnologia de Pernambuco (FACEPE)
LinkOut - more resources
Full Text Sources
Miscellaneous

