Establishment of a New Real-Time Molecular Assay for the Detection of Babanki Virus in Africa
- PMID: 39772151
- PMCID: PMC11680190
- DOI: 10.3390/v16121841
Establishment of a New Real-Time Molecular Assay for the Detection of Babanki Virus in Africa
Abstract
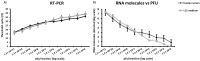
Babanki virus is a subtype of the Sindbis virus, a widespread arthropod-borne alphavirus circulating in Eurasia, Africa, and Oceania. Characterized by rashes and arthritis, clinical infections due to Sindbis were mainly reported in Africa, Australia, Asia, and Europe. However, its sub-type, Babanki virus, was reported in Northern Europe and Africa, where its epidemiology potential remains poorly understood. The diagnosis of alphaviruses is mainly based on serological testing and conventional PCR methods, which have considerable limits. In this study, we developed a real-time qRT-PCR assay for the detection of Babanki virus. The analytical sensitivity and specificity of the newly established assay were evaluated using in vitro standard RNA and related viruses relevant to the African context, respectively. In addition, its diagnostic sensitivity was assessed using a subset of Babanki virus-positive and -negative mosquito pools collected from the field. The new real-time qRT-PCR assay exhibited a 100% specificity, a 95% detection limit of 1 RNA molecule/reaction, and a diagnostic sensitivity of up to 120 pfu/reaction. This newly established assay could be useful not only for the detection of Babanki virus during epidemics but also in future experimental and surveillance studies focusing on their epidemiology and pathogenicity.
Keywords: Africa; Babanki virus; molecular diagnostic; qRT-PCR.
Conflict of interest statement
The authors declare that they have no competing financial interests. The supporting sponsors had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

