Genome Analysis of Anti-Phage Defense Systems and Defense Islands in Stenotrophomonas maltophilia: Preservation and Variability
- PMID: 39772210
- PMCID: PMC11680222
- DOI: 10.3390/v16121903
Genome Analysis of Anti-Phage Defense Systems and Defense Islands in Stenotrophomonas maltophilia: Preservation and Variability
Abstract
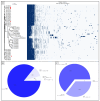
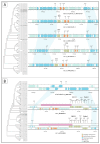
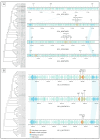
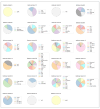
Anti-phage defense systems are widespread in bacteria due to the latter continuous adaptation to infection by bacteriophages (phages). Stenotrophomonas maltophilia has a high degree of intrinsic antibiotic resistance, which makes phage therapy relevant for the treatment of infections caused by this species. Studying the array of anti-phage defense systems that could be found in S. maltophilia helps in better adapting the phages to the systems present in the pathogenic bacteria. Pangenome analysis of the available S. maltophilia strains with complete genomes that were downloaded from GenBank, including five local genomes, indicated a wide set of 72 defense systems and subsystems that varied between the strains. Seven of these systems were present in more than 20% of the studied genomes and the proteins encoded by the systems were variable in most of the cases. A total of 27 defense islands were revealed where defense systems were found; however, more than 60% of the instances of systems were found in four defense islands. Several elements linked to the transfer of these systems were found. No obvious associations between the pattern of distribution of the anti-phage defense systems of S. maltophilia and the phylogenetic features or the isolation site were found.
Keywords: S. maltophilia; anti-phage defense systems; defense islands; phage therapy.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Pompilio A., Crocetta V., Confalone P., Nicoletti M., Petrucca A., Guarnieri S., Fiscarelli E., Savini V., Piccolomini R., Di Bonaventura G. Adhesion to and Biofilm Formation on IB3-1 Bronchial Cells by Stenotrophomonas maltophilia Isolates from Cystic Fibrosis Patients. BMC Microbiol. 2010;10:102. doi: 10.1186/1471-2180-10-102. - DOI - PMC - PubMed
-
- Fluit A.C., Bayjanov J.R., Aguilar M.D., Cantón R., Elborn S., Tunney M.M., Scharringa J., Benaissa-Trouw B.J., Ekkelenkamp M.B. Taxonomic Position, Antibiotic Resistance and Virulence Factor Production by Stenotrophomonas Isolates from Patients with Cystic Fibrosis and Other Chronic Respiratory Infections. BMC Microbiol. 2022;22:129. doi: 10.1186/s12866-022-02466-5. - DOI - PMC - PubMed
-
- WHO|World Health Organization. [(accessed on 7 March 2021)]. Available online: https://www.who.int/health-topics/antimicrobial-resistance.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

