Unique Nucleotide Polymorphism of African Swine Fever Virus Circulating in East Asia and Central Russia
- PMID: 39772214
- PMCID: PMC11680119
- DOI: 10.3390/v16121907
Unique Nucleotide Polymorphism of African Swine Fever Virus Circulating in East Asia and Central Russia
Abstract
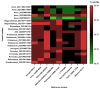
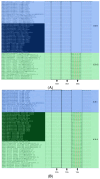
The lack of data on the whole-genome analysis of genotype II African swine fever virus (ASFV) isolates significantly hinders our understanding of its molecular evolution, and as a result, the range of single nucleotide polymorphisms (SNPs) necessary to describe a more accurate and complete scheme of its circulation. In this regard, this study aimed to identify unique SNPs, conduct phylogenetic analysis, and determine the level of homology of isolates obtained in the period from 2019 to 2022 in the central and eastern regions of Russia. Twenty-one whole-genome sequences of genotype II ASFV isolates were assembled, analyzed, and submitted to GenBank. The isolates in eastern Russia form two clades, "Amur 2022" and "Asia". Within the latter clade, five subclusters can be distinguished, each characterized by a unique set of SNPs and indels. The isolates from the central regions of Russia (2019; 2021) form the "Center of Russia" clade, with two subclusters, "Bryansk 2021" and "Center of Russia 2021" (bootstrap confidence index = 99). The presence of the previously unique genetic variant ASFV for the Kaliningrad region in the wild boar population of the Khabarovsk region (eastern Russia; 2021) has also been confirmed.
Keywords: African swine fever virus; Asia; genotype II; molecular epidemiology; single nucleotide polymorphisms.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
Comparative analysis of whole-genome sequences of African swine fever virus (Asfarviridae: Asfivirus) isolates сollected on the territory of the left bank of the Dnieper River in 2023.Vopr Virusol. 2024 Nov 9;69(5):481-494. doi: 10.36233/0507-4088-263. Vopr Virusol. 2024. PMID: 39527770
-
Complete Genome Sequencing and Comparative Phylogenomics of Nine African Swine Fever Virus (ASFV) Isolates of the Virulent East African p72 Genotype IX without Viral Sequence Enrichment.Viruses. 2024 Sep 14;16(9):1466. doi: 10.3390/v16091466. Viruses. 2024. PMID: 39339942 Free PMC article.
-
Spatio-temporal clustering of African swine fever virus (Asfarviridae: Asfivirus) circulating in the Kaliningrad region based on three genome markers.Vopr Virusol. 2024 Jul 5;69(3):241-254. doi: 10.36233/0507-4088-231. Vopr Virusol. 2024. PMID: 38996373
-
African Swine Fever: Disease Dynamics in Wild Boar Experimentally Infected with ASFV Isolates Belonging to Genotype I and II.Viruses. 2019 Sep 13;11(9):852. doi: 10.3390/v11090852. Viruses. 2019. PMID: 31540341 Free PMC article. Review.
-
Pathogenesis of African swine fever in domestic pigs and European wild boar - Lessons learned from recent animal trials.Virus Res. 2019 Oct 2;271:197614. doi: 10.1016/j.virusres.2019.04.001. Epub 2019 Apr 3. Virus Res. 2019. PMID: 30953662 Review.
References
-
- Schulz K., Oļševskis E., Viltrop A., Masiulis M., Staubach C., Nurmoja I., Lamberga K., Seržants M., Malakauskas A., Conraths F.J., et al. Eight Years of African Swine Fever in the Baltic States: Epidemiological Reflections. Pathogens. 2022;11:711. doi: 10.3390/pathogens11060711. - DOI - PMC - PubMed
-
- Beltrán-Alcrudo D., Lubroth J., Depner K., Rocque S. African swine fever in the Caucasus. EMPRES Watch. 2008;1:1–8.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

