DHX15 and Rig-I Coordinate Apoptosis and Innate Immune Signaling by Antiviral RNase L
- PMID: 39772220
- PMCID: PMC11680366
- DOI: 10.3390/v16121913
DHX15 and Rig-I Coordinate Apoptosis and Innate Immune Signaling by Antiviral RNase L
Abstract
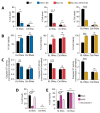
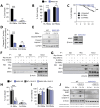
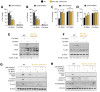
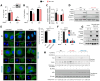
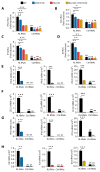
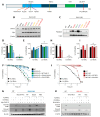
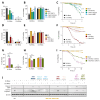
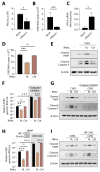
During virus infection, the activation of the antiviral endoribonuclease, ribonuclease L (RNase L), by a unique ligand 2'-5'-oilgoadenylate (2-5A) causes the cleavage of single-stranded viral and cellular RNA targets, restricting protein synthesis, activating stress response pathways, and promoting cell death to establish broad antiviral effects. The immunostimulatory dsRNA cleavage products of RNase L activity (RL RNAs) recruit diverse dsRNA sensors to activate signaling pathways to amplify interferon (IFN) production and activate inflammasome, but the sensors that promote cell death are not known. In this study, we found that DEAH-box polypeptide 15 (DHX15) and retinoic acid-inducible gene I (Rig-I) are essential for apoptosis induced by RL RNAs and require mitochondrial antiviral signaling (MAVS), c-Jun amino terminal kinase (JNK), and p38 mitogen-activated protein kinase (p38 MAPK) for caspase-3-mediated intrinsic apoptosis. In RNase L-activated cells, DHX15 interacts with Rig-I and MAVS, and cells lacking MAVS expression were resistant to apoptosis. RL RNAs induced the transcription of genes for IFN and proinflammatory cytokines by interferon regulatory factor 3 (IRF-3) and nuclear factor kB (NF-kB), while cells lacking both DHX15 and Rig-I showed a reduced induction of cytokines. However, apoptotic cell death is independent of both IRF-3 and NF-kB, suggesting that cytokine and cell death induction by RL RNAs are uncoupled. The RNA binding of both DHX15 and Rig-I is required for apoptosis induction, and the expression of both single proteins in cells lacking both DHX15 and Rig-I is insufficient to promote cell death by RL RNAs. Cell death induced by RL RNAs suppressed Coxsackievirus B3 (CVB3) replication, and inhibiting caspase-3 activity or cells lacking IRF-3 showed that the induction of apoptosis directly resulted in the CVB3 antiviral effect, and the effects were independent of the role of IRF-3.
Keywords: DHX15; RNase L; Rig-I; apoptosis; innate immune signaling.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

