A Global Collaborative Comparison of SARS-CoV-2 Antigenicity Across 15 Laboratories
- PMID: 39772242
- PMCID: PMC11680265
- DOI: 10.3390/v16121936
A Global Collaborative Comparison of SARS-CoV-2 Antigenicity Across 15 Laboratories
Abstract
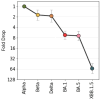
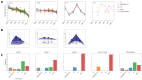
Setting up a global SARS-CoV-2 surveillance system requires an understanding of how virus isolation and propagation practices, use of animal or human sera, and different neutralisation assay platforms influence assessment of SARS-CoV-2 antigenicity. In this study, with the contribution of 15 independent laboratories across all WHO regions, we carried out a controlled analysis of neutralisation assay platforms using the first WHO International Standard for antibodies to SARS-CoV-2 variants of concern (source: NIBSC). Live virus isolates (source: WHO BioHub or individual labs) or spike plasmids (individual labs) for pseudovirus production were used to perform neutralisation assays using the same serum panels. When comparing fold drops, excellent data consistency was observed across the labs using common reagents, including between pseudovirus and live virus neutralisation assays (RMSD of data from mean fold drop was 0.59). Utilising a Bayesian model, geometric mean titres and assay titre magnitudes (offsets) can describe the data efficiently. Titre magnitudes were seen to vary largely even for labs within the same assay group. We have observed that overall, live Microneutralisation assays tend to have the lowest titres, whereas Pseudovirus Neutralisation have the highest (with a mean difference of 3.2 log2 units between the two). These findings are relevant for laboratory networks, such as the WHO Coronavirus Laboratory Network (CoViNet), that seek to support a global surveillance system for evolution and antigenic characterisation of variants to support monitoring of population immunity and vaccine composition policy.
Keywords: Bayesian model; COVID-19; SARS-CoV-2; antigenicity; global surveillance; neutralisation.
Conflict of interest statement
I.O.D., J.O.A., F.A.S., T.H. (Taweewun Hunsawong), M.B. (Matthias Budt), M.M.S. (Melanie M. Schmitt), C.D., B.M., L.F.L.T., M.F.d.A., A.C.A.d.O., P.A.N., P.D.B.C.C., M.M.S. (Marilda Mendonça Siqueira), J.-M.G., M.B. (Meriem Bekliz), T.W., S.T., B.L.H., A.Z.M., D.S., S.M.S.C., J.K.C.L., L.L.M.P., M.P., N.L., J.O.A., I.O.O., D.Z., O.E., I.H.-G., T.H. (Tandile Hermanus), K.R., J.L., J.N.B., C.C., S.I.R., P.D., T.H. (Tarteel Hassan), J.C., M.D.V.K., S.T., P.B., G.M., D.S. and L.S. declare no competing interests I.E. has received research funding for an unrelated project (IIT) and speaker and consultancy fees from Moderna. V.M.C. is a co-inventor on a patent application entitled “Methods and reagents for diagnosis of SARS-CoV-2 infection” (patent application no eP3809137a1). J.K. is listed as inventor on patents related to VSV-based oncolytic viruses. A.S. has received an honorarium from Pfizer for consulting.
Figures


References
-
- Chatterjee S., Bhattacharya M., Nag S., Dhama K., Chakraborty C. A Detailed Overview of SARS-CoV-2 Omicron: Its Sub-Variants, Mutations and Pathophysiology, Clinical Characteristics, Immunological Landscape, Immune Escape, and Therapies. Viruses. 2023;15:167. doi: 10.3390/v15010167. - DOI - PMC - PubMed
-
- Subissi L., Otieno J.R., Worp N., Attar Cohen H., Oude Munnink B.B., Abu-Raddad L.J., Alm E., Barakat A., Barclay W.S., Bhiman J.N., et al. An updated framework for SARS-CoV-2 variants reflects the unpredictability of viral evolution. Nat. Med. 2024;30:1–4. doi: 10.1038/s41591-024-02949-0. - DOI - PubMed
-
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

