Respiratory Virus-Specific and Time-Dependent Interference of Adenovirus Type 2, SARS-CoV-2 and Influenza Virus H1N1pdm09 During Viral Dual Co-Infection and Superinfection In Vitro
- PMID: 39772252
- PMCID: PMC11680187
- DOI: 10.3390/v16121947
Respiratory Virus-Specific and Time-Dependent Interference of Adenovirus Type 2, SARS-CoV-2 and Influenza Virus H1N1pdm09 During Viral Dual Co-Infection and Superinfection In Vitro
Abstract
Background: Understanding the interference patterns of respiratory viruses could be important for shedding light on potential strategies to combat these human infectious agents.
Objective: To investigate the possible interactions between adenovirus type 2 (AdV2), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and influenza A/H1N1 pandemic (H1N1pdm09) using the A549 cell line.
Methods: Single infections, co-infections, and superinfections (at 3 and 24 h after the first virus infection) were performed by varying the multiplicity of infection (MOI). Virus replication kinetics and the mRNA expression of IFN-α, IL-1α and IL-6 were assessed by real-time qPCR.
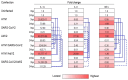
Results: Co-infection experiments showed different growth dynamics, depending on the presence of the specific virus and time. AdV2 replication remained stable or possibly enhanced in the presence of co-infection with each of the two H1N1pdm09 and SARS-CoV-2 viruses used. In contrast, SARS-CoV-2 replication was facilitated by H1N1pdm09 but hindered by AdV2, indicating possible different interactions. Finally, H1N1pdm09 replication exhibited variably effectiveness in the presence of AdV2 and SARS-CoV-2. Superinfection experiments showed that the replication of all viruses was affected by time and MOI. The mRNA expression of IFN-α, IL-1α and IL-6 showed divergent results depending on the virus used and the time of infection.
Conclusions: Further investigation of co-infection or superinfection may be helpful in understanding the potential relationship involved in the outcome of viral respiratory infection in the human population.
Keywords: SARS-CoV-2; adenovirus; co-infection; influenza virus; superinfection.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Zhang A.J., Lee A.C., Chan J.F., Liu F., Li C., Chen Y., Chu H., Lau S.Y., Wang P., Chan C.C., et al. Coinfection by severe acute respiratory syndrome coronavirus 2 and influenza A(H1N1)pdm09 virus enhances the severity of pneumonia in golden syrian hamsters. Clin. Infect. Dis. 2021;72:e978–e992. doi: 10.1093/cid/ciaa1747. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

