A hTfR1 Receptor-Specific VHH Antibody Neutralizes Pseudoviruses Expressing Glycoproteins from Junín and Machupo Viruses
- PMID: 39772257
- PMCID: PMC11680233
- DOI: 10.3390/v16121951
A hTfR1 Receptor-Specific VHH Antibody Neutralizes Pseudoviruses Expressing Glycoproteins from Junín and Machupo Viruses
Abstract
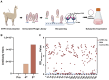
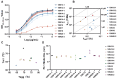
The Junín virus (JUNV) is one of the New World arenaviruses that cause severe hemorrhagic fever. Human transferrin receptor 1 (hTfR1) has been identified as the main receptor for JUNV for virus entry into host cells. To date, no treatment has been approved for JUNV. Herein, we investigated 12 anti-hTfR1 VHH (variable domain of the heavy chain of heavy-chain antibody) antibodies and confirmed their interaction with hTfR1. Most of them could bind to the hTfR1 apical domain, which is the glycoprotein 1 (GP1) binding domain of JUNV. Among them, 18N18 exhibited neutralizing activity against both the human immunodeficiency virus (HIV)-vectored lentiviral Junín pseudoviruses and the recombinant vesicular stomatitis virus (VSV)-vectored Junín pseudoviruses. We also verified that 18N18 blocked the interaction between hTfR1 and JUNV GP1. In addition, 18N18 could neutralize another New World arenavirus, the Machupo virus. Using AlphaFold 3-based simulation of 18N18-hTfR1 docking, we determined that 18N18's binding epitope was located at the JUNV GP1 binding epitope. 18N18 represents a candidate for JUNV treatment and provides a potential approach that could be applied to New World arenaviruses.
Keywords: Junín viruses; Machupo viruses; VHH antibody; transferrin receptor 1.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

