Viral Oncogenesis: Synergistic Role of Genome Integration and Persistence
- PMID: 39772271
- PMCID: PMC11728759
- DOI: 10.3390/v16121965
Viral Oncogenesis: Synergistic Role of Genome Integration and Persistence
Abstract
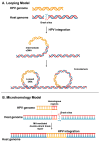
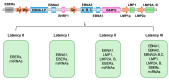
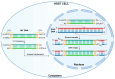
Persistence is a strategy used by many viruses to evade eradication by the immune system, ensuring their permanence and transmission within the host and optimizing viral fitness. During persistence, viruses can trigger various phenomena, including target organ damage, mainly due to an inflammatory state induced by infection, as well as cell proliferation and/or immortalization. In addition to immune evasion and chronic inflammation, factors contributing to viral persistence include low-level viral replication, the accumulation of viral mutants, and, most importantly, maintenance of the viral genome and reliance on viral oncoprotein production. This review focuses on the process of genome integration, which may occur at different stages of infection (e.g., HBV), during the chronic phase of infection (e.g., HPV, EBV), or as an essential part of the viral life cycle, as seen in retroviruses (HIV, HTLV-1). It also explores the close relationship between integration, persistence, and oncogenesis. Several models have been proposed to describe the genome integration process, including non-homologous recombination, looping, and microhomology models. Integration can occur either randomly or at specific genomic sites, often leading to genome destabilization. In some cases, integration results in the loss of genomic regions or impairs the regulation of oncogene and/or oncosuppressor expression, contributing to tumor development.
Keywords: Epstein–Barr virus; episome; genome integration; hepatitis B virus; latency; oncogenic viruses; papillomavirus; persistence; retrovirus.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Boldogh I., Albrecht T., Porter D.D. Persistent Viral Infections. In: Baron S., editor. Medical Microbiology. University of Texas Medical Branch at Galveston; Galveston, TX, USA: 1996. - PubMed
-
- Crawford D.H. Viruses: A Very Short Introduction. Oxford University Press; Oxford, UK: 2018. Persistent Viruses; pp. 75–91.
-
- Crawford D.H. Viruses: A Very Short Introduction. Oxford University Press; Oxford, UK: 2018. Tumour Viruses; pp. 92–109.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

