2-Cyanopyrimidine-Containing Molecules for N-Terminal Selective Cyclization of Phage-Displayed Peptides
- PMID: 39772425
- PMCID: PMC11744668
- DOI: 10.1021/acschembio.4c00725
2-Cyanopyrimidine-Containing Molecules for N-Terminal Selective Cyclization of Phage-Displayed Peptides
Abstract
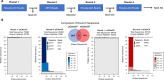
Current methods for the macrocyclization of phage-displayed peptides often rely on small molecule linkers that nonspecifically react with targeted amino acid residues. To expand tool kits for more regioselective macrocyclization of phage-displayed peptides, this study explores the unique condensation reaction between an N-terminal cysteine and nitrile along with the reactivity of an internal cysteine. Five 2-cyanopyrimidine derivatives were synthesized for this purpose and evaluated for their selective macrocyclization of a protein-fused model peptide. Among these, two novel linkers, 2-chloro-N-(2-cyanopyrimidin-5-yl)acetamide (pCAmCP) and 2-chloro-N-(2-cyanopyrimidin-4-yl)acetamide (mCAmCP), emerged as efficient molecules and were demonstrated to macrocyclize phage-displayed peptide libraries flanked by an N-terminal and an internal cysteine. Using these linkers to generate macrocyclic peptide libraries displayed on phages, peptide ligands for the ZNRF3 extracellular domain were successfully identified. One of the identified peptides, Z27S1, exhibited potent binding to ZNRF3 with a KD value of 360 nM. Notably, the selection results revealed distinct peptide enrichment patterns depending on whether mCAmCP or pCAmCP was used, underscoring the significant impact of linker choice on macrocyclic peptide identification. Overall, this study validates the development of two novel regioselective, small molecule linkers for phage display of macrocyclic peptides and highlights the benefits of employing multiple linkers during phage selections.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



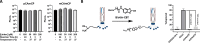

Similar articles
-
Proximity-driven site-specific cyclization of phage-displayed peptides.Nat Commun. 2024 Aug 24;15(1):7308. doi: 10.1038/s41467-024-51610-4. Nat Commun. 2024. PMID: 39181880 Free PMC article.
-
Homodimeric peptides displayed by the major coat protein of filamentous phage.J Mol Biol. 2000 Jul 7;300(2):307-20. doi: 10.1006/jmbi.2000.3850. J Mol Biol. 2000. PMID: 10873467
-
A Genetically Encoded, Phage-Displayed Cyclic-Peptide Library.Angew Chem Int Ed Engl. 2019 Oct 28;58(44):15904-15909. doi: 10.1002/anie.201908713. Epub 2019 Sep 9. Angew Chem Int Ed Engl. 2019. PMID: 31398275 Free PMC article. Review.
-
Selection of Peptide-Bismuth Bicycles Using Phage Display.ACS Chem Biol. 2024 May 17;19(5):1040-1044. doi: 10.1021/acschembio.4c00099. Epub 2024 Apr 15. ACS Chem Biol. 2024. PMID: 38620022
-
Biosynthetic Strategies for Macrocyclic Peptides.Molecules. 2021 Jun 1;26(11):3338. doi: 10.3390/molecules26113338. Molecules. 2021. PMID: 34206124 Free PMC article. Review.
References
-
- Lau J.; Bloch P.; Schäffer L.; Pettersson I.; Spetzler J.; Kofoed J.; Madsen K.; Knudsen L. B.; McGuire J.; Steensgaard D. B.; Strauss H. M.; Gram D. X.; Knudsen S. M.; Nielsen F. S.; Thygesen P.; Reedtz-Runge S.; Kruse T. Discovery of the Once-Weekly Glucagon-Like Peptide-1 (GLP-1) Analogue Semaglutide. J. Med. Chem. 2015, 58 (18), 7370–7380. 10.1021/acs.jmedchem.5b00726. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

