Regulation of Cell-Nanoparticle Interactions through Mechanobiology
- PMID: 39772635
- PMCID: PMC11849000
- DOI: 10.1021/acs.nanolett.4c04290
Regulation of Cell-Nanoparticle Interactions through Mechanobiology
Abstract
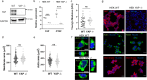
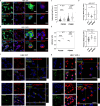
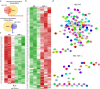
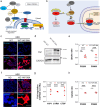
Bio-nano interactions have been extensively explored in nanomedicine to develop selective delivery strategies and reduce systemic toxicity. To enhance the delivery of nanocarriers to cancer cells and improve the therapeutic efficiency, different nanomaterials have been developed. However, the limited clinical translation of nanoparticle-based therapies, largely due to issues associated with poor targeting, requires a deeper understanding of the biological phenomena underlying cell-nanoparticle interactions. In this context, we investigate the molecular and cellular mechanobiology parameters that control such interactions. We demonstrate that the pharmacological inhibition or the genetic ablation of the key mechanosensitive component of the Hippo pathway, i.e., yes-associated protein, enhances nanoparticle internalization by 1.5-fold. Importantly, this phenomenon occurs independently of nanoparticle properties, such as size, or cell properties such as surface area and stiffness. Our study reveals that the internalization of nanoparticles in target cells can be controlled by modulating cell mechanosensing pathways, potentially enhancing nanotherapy specificity.
Keywords: bio−nano interactions; mechanobiology; mechanotransduction; nanoparticles.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
-
- Wilhelm S.; Tavares A. J.; Dai Q.; Ohta S.; Audet J.; Dvorak H. F.; Chan W. C. W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016, 1 (5), 16014.10.1038/natrevmats.2016.14. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

