Subthreshold repetitive transcranial magnetic stimulation induces cortical layer-, brain region-, and protocol-dependent neural plasticity
- PMID: 39772671
- PMCID: PMC11708880
- DOI: 10.1126/sciadv.ado6705
Subthreshold repetitive transcranial magnetic stimulation induces cortical layer-, brain region-, and protocol-dependent neural plasticity
Abstract
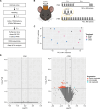
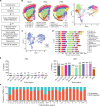
Repetitive transcranial magnetic stimulation (rTMS) is commonly used to study the brain or as a treatment for neurological disorders, but the neural circuits and molecular mechanisms it affects remain unclear. To determine the molecular mechanisms of rTMS and the brain regions they occur in, we used spatial transcriptomics to map changes to gene expression across the mouse brain in response to two commonly used rTMS protocols. Our results revealed that rTMS alters the expression of genes related to several cellular processes and neural plasticity mechanisms across the brain, which was both brain region- and rTMS protocol-dependent. In the cortex, the effect of rTMS was dependent not only on the cortical region but also on each cortical layer. These findings uncover the diverse molecular mechanisms induced by rTMS, which will be useful in interpreting its effects on cortical and subcortical circuits.
Figures





References
-
- Lenz M., Platschek S., Priesemann V., Becker D., Willems L. M., Ziemann U., Deller T., Müller-Dahlhaus F., Jedlicka P., Vlachos A., Repetitive magnetic stimulation induces plasticity of excitatory postsynapses on proximal dendrites of cultured mouse CA1 pyramidal neurons. Brain Struct. Funct. 220, 3323–3337 (2015). - PubMed
-
- Tang A. D., Bennett W., Bindoff A. D., Bolland S., Collins J., Langley R. C., Garry M. I., Summers J. J., Hinder M. R., Rodger J., Canty A. J., Subthreshold repetitive transcranial magnetic stimulation drives structural synaptic plasticity in the young and aged motor cortex. Brain Stimul. 14, 1498–1507 (2021). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

