Non-adapted bacterial infection suppresses plant reproduction
- PMID: 39772678
- PMCID: PMC11708875
- DOI: 10.1126/sciadv.ads7738
Non-adapted bacterial infection suppresses plant reproduction
Abstract
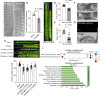
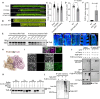
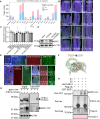
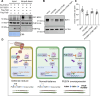
Environmental stressors, including pathogens, substantially affect the growth of host plants. However, how non-adapted bacteria influence nonhost plants has not been reported. Here, we reveal that infection of Arabidopsis flowers by Xanthomonas oryzae pv. oryzae PXO99A, a bacterial pathogen causing rice blight disease, suppresses ovule initiation and reduces seed number without causing visible disease symptoms. TleB, secreted by the type VI secretion system (T6SS), interacts with plant E3 ligase PUB14 and disrupts the function of the PUB14-BZR1 module, leading to decreased ovule initiation and seed yield. On the other site, PUB14 concurrently promotes TleB's degradation. Our findings indicate that bacterial infections in nonhost plants directly repress offspring production. The regulatory mechanism by effectors PUB14-BZR1 is widely present, suggesting that plants may balance reproduction and defense and produce fewer offspring to conserve resources, thus enabling them to remain in a standby mode prepared for enhanced resistance.
Figures

References
-
- Jones J. D., Dangl J. L., The plant immune system. Nature 444, 323–329 (2006). - PubMed
-
- Senthil-Kumar M., Mysore K. S., Nonhost resistance against bacterial pnaturathogens: Retrospectives and prospects. Annu. Rev. Phytopathol. 51, 407–427 (2013). - PubMed
-
- Wang Y. T., Chen H. W., Shao M. S., Zhu T., Li S. S., Olsson P. A., Hammer E. C., Arbuscular mycorrhizal fungi trigger danger-associated peptide signaling and inhibit carbon-phosphorus exchange with nonhost plants. Plant Cell Environ. 46, 2206–2221 (2023). - PubMed
-
- He Z., Webster S., He S. Y., Growth-defense trade-offs in plants. Curr. Biol. 32, R634–R639 (2022). - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

