Infection-induced lysine lactylation enables herpesvirus immune evasion
- PMID: 39772686
- PMCID: PMC11708889
- DOI: 10.1126/sciadv.ads6215
Infection-induced lysine lactylation enables herpesvirus immune evasion
Abstract
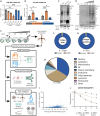
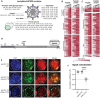
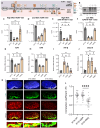
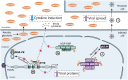
Aerobic glycolysis is a hallmark of many viral infections, leading to substantial accumulation of lactate. However, the regulatory roles of lactate during viral infections remain poorly understood. Here, we report that human cytomegalovirus (HCMV) infection leverages lactate to induce widespread protein lactylation and promote viral spread. We establish that lactyllysine is enriched in intrinsically disordered regions, regulating viral protein condensates and immune signaling transduction. Dynamic lactylation of immune factors suppresses immunity, a feature we show to be shared for HCMV and herpes simplex virus 1 infections, through regulation of RNA binding protein 14 and interferon-γ-inducible protein 16 (IFI16). K90 lactylation of the viral DNA sensor IFI16 inhibits recruitment of the DNA damage response kinase DNA-PK, preventing IFI16-driven virus gene repression and cytokine induction. Together, we characterize global protein lactylation dynamics during virus infection, finding that virus-induced lactate contributes to its immune evasion through direct inhibition of immune signaling pathways.
Figures




References
-
- Certo M., Tsai C. H., Pucino V., Ho P. C., Mauro C., Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol. 21, 151–161 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

