Prediction of protein biophysical traits from limited data: a case study on nanobody thermostability through NanoMelt
- PMID: 39772905
- PMCID: PMC11730357
- DOI: 10.1080/19420862.2024.2442750
Prediction of protein biophysical traits from limited data: a case study on nanobody thermostability through NanoMelt
Abstract
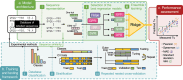
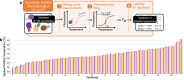
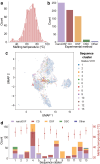
In-silico prediction of protein biophysical traits is often hindered by the limited availability of experimental data and their heterogeneity. Training on limited data can lead to overfitting and poor generalizability to sequences distant from those in the training set. Additionally, inadequate use of scarce and disparate data can introduce biases during evaluation, leading to unreliable model performances being reported. Here, we present a comprehensive study exploring various approaches for protein fitness prediction from limited data, leveraging pre-trained embeddings, repeated stratified nested cross-validation, and ensemble learning to ensure an unbiased assessment of the performances. We applied our framework to introduce NanoMelt, a predictor of nanobody thermostability trained with a dataset of 640 measurements of apparent melting temperature, obtained by integrating data from the literature with 129 new measurements from this study. We find that an ensemble model stacking multiple regression using diverse sequence embeddings achieves state-of-the-art accuracy in predicting nanobody thermostability. We further demonstrate NanoMelt's potential to streamline nanobody development by guiding the selection of highly stable nanobodies. We make the curated dataset of nanobody thermostability freely available and NanoMelt accessible as a downloadable software and webserver.
Keywords: Biological sciences – biophysics and computational biology; Protein fitness; antibody design; antibody engineering; ensemble model; machine learning; nanobody; semi-supervised learning; thermostability.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Kim JY, Yoo H-W, Lee P-G, Lee S-G, Seo J-H, Kim B-G.. In vivo protein evolution, next generation protein engineering strategy: from random approach to target-specific approach. Biotechnol Bioprocess Eng. 2019;24(1):85–94. doi: 10.1007/s12257-018-0394-2. - DOI
-
- Meier J, Rao R R, Verkuil R, Liu J, Sercu T, Rives A.. Language models enable zero-shot prediction of the effects of mutations on protein function in. In: Ranzato M, Beygelzimer A, Dauphin Y, Liang PS, Wortman Vaughan J, editors. Advances in neural information processing systems. Red Hook, NY, USA: Curran Associates, Inc.; 2021. p. 29287–29303.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
