PLK2 disrupts autophagic flux to promote SNCA/α-synuclein pathology
- PMID: 39773002
- PMCID: PMC12283002
- DOI: 10.1080/15548627.2024.2448914
PLK2 disrupts autophagic flux to promote SNCA/α-synuclein pathology
Abstract
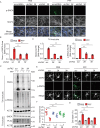
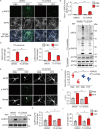
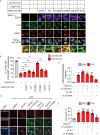
The aggregation and transmission of SNCA/α-synuclein (synuclein, alpha) is a hallmark pathology of Parkinson disease (PD). PLK2 (polo like kinase 2) is an evolutionarily conserved serine/threonine kinase that is more abundant in the brains of all family members, is highly expressed in PD, and is linked to SNCA deposition. However, in addition to its role in phosphorylating SNCA, the role of PLK2 in PD and the mechanisms involved in triggering neurodegeneration remain unclear. Here, we found that PLK2 regulated SNCA pathology independently of S129. Overexpression of PLK2 promoted SNCA preformed fibril (PFF)-induced aggregation of wild-type SNCA and mutant SNCAS129A. Genetic or pharmacological inhibition of PLK2 attenuated SNCA deposition and neurotoxicity. Mechanistically, PLK2 exacerbated the propagation of SNCA pathology by impeding the clearance of SNCA aggregates by blocking macroautophagic/autophagic flux. We further showed that PLK2 phosphorylated S1098 of DCTN1 (dynactin 1), a protein that controls the movement of organelles, leading to impaired autophagosome-lysosome fusion. Furthermore, genetic suppression of PLK2 alleviated SNCA aggregation and motor dysfunction in vivo. Our findings suggest that PLK2 negatively regulates autophagy, promoting SNCA pathology, suggesting a role for PLK2 in PD.Abbreviation: AD: Alzheimer disease; AMPK: AMP-activated protein kinase; CASP3: caspase 3; DCTN1: dynactin 1; LBs: lewy bodies; LDH: lactate dehydrogenase; LAMP1: lysosomal associated membrane protein 1; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MAP2: microtubule associated protein 2; MTOR: mechanistic target of rapamycin kinase; NH4Cl: ammonium chloride; p-SNCA: phosphorylation of SNCA at S129; PD: Parkinson disease; PFF: preformed fibril; PI: propidium iodide; PLK2: polo like kinase 2; PRKAA/AMPK: protein kinase AMP-activated catalytic subunit alpha; shRNA: short hairpin RNA; SNCA: synuclein, alpha; SQSTM1/p62: sequestosome 1; TH: tyrosine hydroxylase; TX: Triton X-100; ULK1: unc-51 like autophagy activating kinase 1.
Keywords: Autophagic flux; DCTN1; PLK2; Parkinson disease; autophagosome-lysosome fusion.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
