Multi‑omics identification of a novel signature for serous ovarian carcinoma in the context of 3P medicine and based on twelve programmed cell death patterns: a multi-cohort machine learning study
- PMID: 39773329
- PMCID: PMC11707953
- DOI: 10.1186/s10020-024-01036-x
Multi‑omics identification of a novel signature for serous ovarian carcinoma in the context of 3P medicine and based on twelve programmed cell death patterns: a multi-cohort machine learning study
Abstract
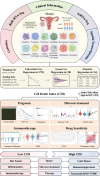
Background: Predictive, preventive, and personalized medicine (PPPM/3PM) is a strategy aimed at improving the prognosis of cancer, and programmed cell death (PCD) is increasingly recognized as a potential target in cancer therapy and prognosis. However, a PCD-based predictive model for serous ovarian carcinoma (SOC) is lacking. In the present study, we aimed to establish a cell death index (CDI)-based model using PCD-related genes.
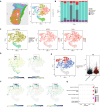
Methods: We included 1254 genes from 12 PCD patterns in our analysis. Differentially expressed genes (DEGs) from the Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) were screened. Subsequently, 14 PCD-related genes were included in the PCD-gene-based CDI model. Genomics, single-cell transcriptomes, bulk transcriptomes, spatial transcriptomes, and clinical information from TCGA-OV, GSE26193, GSE63885, and GSE140082 were collected and analyzed to verify the prediction model.
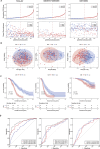
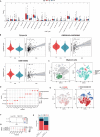
Results: The CDI was recognized as an independent prognostic risk factor for patients with SOC. Patients with SOC and a high CDI had lower survival rates and poorer prognoses than those with a low CDI. Specific clinical parameters and the CDI were combined to establish a nomogram that accurately assessed patient survival. We used the PCD-genes model to observe differences between high and low CDI groups. The results showed that patients with SOC and a high CDI showed immunosuppression and hardly benefited from immunotherapy; therefore, trametinib_1372 and BMS-754807 may be potential therapeutic agents for these patients.
Conclusions: The CDI-based model, which was established using 14 PCD-related genes, accurately predicted the tumor microenvironment, immunotherapy response, and drug sensitivity of patients with SOC. Thus this model may help improve the diagnostic and therapeutic efficacy of PPPM.
Keywords: Cell death index; Predictive model; Predictive preventive and personalized medicine (PPPM/3PM); Programmed cell death; Serous ovarian carcinoma.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The TCGA and GEO databases are publicly available. Users can download the data for free for research purposes and publish related articles. The authors declare no ethical issues or conflicts of interest. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

