Multiple introductions of NRCS-A Staphylococcus capitis to the neonatal intensive care unit drive neonatal bloodstream infections: a case-control and environmental genomic survey
- PMID: 39773387
- PMCID: PMC11706212
- DOI: 10.1099/mgen.0.001340
Multiple introductions of NRCS-A Staphylococcus capitis to the neonatal intensive care unit drive neonatal bloodstream infections: a case-control and environmental genomic survey
Abstract
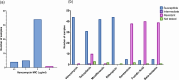
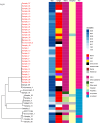
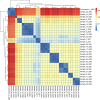
Background. The Staphylococcus capitis NRCS-A strain has emerged as a global cause of late-onset sepsis associated with outbreaks in neonatal intensive care units (NICUs) whose transmission is incompletely understood.Methods. Demographic and clinical data for 45 neonates with S. capitis and 90 with other coagulase-negative staphylococci (CoNS) isolated from sterile sites were reviewed, and clinical significance was determined. S. capitis isolated from 27 neonates at 2 hospitals between 2017 and 2022 underwent long-read (ONT) (n=27) and short-read (Illumina) sequencing (n=18). These sequences were compared with S. capitis sequenced from blood culture isolates from other adult and paediatric patients in the same hospitals (n=6), S. capitis isolated from surface swabs (found in 5/150 samples), rectal swabs (in 2/69 samples) in NICU patients and NICU environmental samples (in 5/114 samples). Reads from all samples were mapped to a hybrid assembly of a local sterile site strain, forming a complete UK NRCS-A reference genome, for outbreak analysis and comparison with 826 other S. capitis from the UK and Germany.Results. S. capitis bacteraemia was associated with increased length of NICU stay at sampling (median day 22 vs day 12 for other CoNS isolated; P=0.05). A phylogeny of sequenced S. capitis revealed a cluster comprised of 25/27 neonatal sterile site isolates and 3/5 superficial, 2/2 rectal and 1/5 environmental isolates. No isolates from other wards belonged to this cluster. Phylogenetic comparison with published sequences confirmed that the cluster was NRCS-A outbreak strain but found a relatively high genomic diversity (mean pairwise distance of 84.9 SNPs) and an estimated NRCS-A S. capitis molecular clock of 5.1 SNPs/genome/year (95% credibility interval 4.3-5.9). The presence of S. capitis in superficial cultures did not correlate with neonatal bacteraemia, but both neonates with rectal NRCS-A S. capitis carriage identified also experienced S. capitis bacteraemia.Conclusions. S. capitis bacteraemia occurred in patients with longer NICU admission than other CoNS. Genomic analysis confirms clinically significant infections with the NRCS-A S. capitis strain, distinct from non-NICU clinical samples. Multiple introductions of S. capitis, rather than prolonged environmental persistence, were seen over 5 years of infections.
Keywords: Staphylococcus capitis; antimicrobial resistance; infection control; late-onset sepsis; neonates.
Conflict of interest statement
The authors declare there are no conflicts of interest.
Figures
Similar articles
-
Genomic analysis of Staphylococcus capitis isolated from blood cultures in neonates at a neonatal intensive care unit in Sweden.Eur J Clin Microbiol Infect Dis. 2019 Nov;38(11):2069-2075. doi: 10.1007/s10096-019-03647-3. Epub 2019 Aug 9. Eur J Clin Microbiol Infect Dis. 2019. PMID: 31396832 Free PMC article.
-
Characterisation of neonatal Staphylococcus capitis NRCS-A isolates compared with non NRCS-A Staphylococcus capitis from neonates and adults.Microb Genom. 2023 Oct;9(10):001106. doi: 10.1099/mgen.0.001106. Microb Genom. 2023. PMID: 37791541 Free PMC article.
-
Genomic Analysis of Multiresistant Staphylococcus capitis Associated with Neonatal Sepsis.Antimicrob Agents Chemother. 2018 Oct 24;62(11):e00898-18. doi: 10.1128/AAC.00898-18. Print 2018 Nov. Antimicrob Agents Chemother. 2018. PMID: 30150477 Free PMC article.
-
Staphylococcus capitis and NRCS-A clone: the story of an unrecognized pathogen in neonatal intensive care units.Clin Microbiol Infect. 2019 Sep;25(9):1081-1085. doi: 10.1016/j.cmi.2019.03.009. Epub 2019 Mar 27. Clin Microbiol Infect. 2019. PMID: 30928561 Review.
-
Staphylococcus capitis: Review of Its Role in Infections and Outbreaks.Antibiotics (Basel). 2023 Mar 29;12(4):669. doi: 10.3390/antibiotics12040669. Antibiotics (Basel). 2023. PMID: 37107031 Free PMC article. Review.
References
-
- Perin J, Mulick A, Yeung D, Villavicencio F, Lopez G, et al. Global, regional, and national causes of under-5 mortality in 2000-19: an updated systematic analysis with implications for the Sustainable Development Goals. Lancet Child Adolesc Health . 2022;6:106–115. doi: 10.1016/S2352-4642(21)00311-4. - DOI - PMC - PubMed
-
- British Association of Perinatal Medicine Perinatal management of extreme preterm birth before weeks of gestation: a framework for practice. https://www.bapm.org/resources/80-perinatal-management-of-extreme-preter... n.d. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

