Differential impact of TIM-3 ligands on NK cell function
- PMID: 39773563
- PMCID: PMC11748930
- DOI: 10.1136/jitc-2024-010618
Differential impact of TIM-3 ligands on NK cell function
Abstract
Background: The transmembrane protein T-cell immunoglobulin and mucin-domain containing molecule 3 (TIM-3) is an immune checkpoint receptor that is expressed by a variety of leukocyte subsets, particularly in the tumor microenvironment. An effective TIM-3-targeting therapy should account for multiple biological factors, including the disease setting, the specific cell types involved and their varying sensitivities to the four putative TIM-3 ligands (galectin-9, phosphatidylserine, high mobility group protein B1 and carcinoembryonic antigen cell adhesion molecule 1), each of which engages a unique binding site on the receptor's variable immunoglobulin domain. The primary objectives of this study were to assess the prevalence and function of TIM-3+ natural killer (NK) cells in patients with head and neck squamous cell carcinoma (HNSCC), determine whether the four TIM-3 ligands differentially affect TIM-3+ NK cell functions, identify the most immunosuppressive ligand, and evaluate whether targeting ligand-mediated TIM-3 signaling enhances NK cell effector functions.
Methods: Single-cell RNA sequencing and flow cytometry were used to study the prevalence, phenotypes and function of TIM-3+ NK cells in HNSCC patient tumors and blood. In vitro killing, proliferation and cytokine production assays were implemented to evaluate whether the four TIM-3 ligands differentially modulate TIM-3+ NK cell functions, and whether disruption of TIM-3/ligand interaction can enhance NK cell-mediated antitumor effector mechanisms. Finally, The Cancer Genome Atlas survival analysis and digital spatial profiling were employed to study the potential impact of etiology-associated differences on patients with HNSCC outcomes.
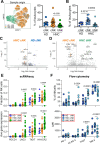
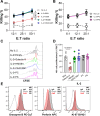
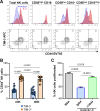
Results: We demonstrate that TIM-3 is highly prevalent on circulating and tumor-infiltrating NK cells. It co-expresses with CD44 and marks NK cells with heightened effector potential. Among the four putative TIM-3 ligands, galectin-9 most consistently suppresses NK cell-mediated cytotoxicity and proliferation through TIM-3 and CD44 signaling, respectively, but promotes IFN-γ release in a TIM-3-dependent manner. Among patients with HNSCC, an elevated intratumoral TIM-3+ NK cell gene signature associates with worse outcomes, specifically in those with human papillomavirus (HPV)+ disease, potentially attributable to higher galectin-9 levels in HPV+ versus HPV- patients.
Conclusions: Our findings underscore the complex functional impact of TIM-3 ligand signaling, which is consistent with recent clinical trials suggesting that targeting TIM-3 alone is suboptimal as an immunotherapeutic approach for treating malignancies.
Keywords: Head and Neck Cancer; Immune Checkpoint Inhibitor; Innate; Natural killer - NK; Tumor Microenvironment.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: RLF is co-founder and stockholder of Novasenta, and declares competing interests with Aduro Biotech (consulting), AstraZeneca/MedImmune (clinical trial, research funding), Bristol Myers Squibb (advisory board, clinical trial, research funding), EMD Serono (advisory board), MacroGenics (advisory board), Merck (advisory board, clinical trial), Novasenta (consulting, stock, research funding), Numab Therapeutics AG (advisory board), Pfizer (advisory board), Sanofi (consultant), Tesaro (research funding) and Zymeworks (consultant). TCB serves on the scientific advisory board of Walking Fish Therapeutics, Bespoke Therapeutics, Mestag Therapeutics, Kalivir Therapeutics, and Galvanize Therapeutics. TCB is a consultant for Attivare Therapeutics and Tabby Therapeutics. LV declares US Patent Number: 10,543,264, a methodology licensed to INmune Bio where DN-TNF can be used to prevent or treat malignancies. RB declares PCT/US15/612657 (Cancer Immunotherapy), PCT/US18/36052 (Microbiome Biomarkers for Anti-PD-1/PD-L1 Responsiveness: Diagnostic, Prognostic and Therapeutic Uses Thereof), PCT/US63/055227 (Methods and Compositions for Treating Autoimmune and Allergic Disorders).
Figures




References
-
- Zeidan AM, Ando K, Rauzy O, et al. Sabatolimab plus hypomethylating agents in previously untreated patients with higher-risk myelodysplastic syndromes (STIMULUS-MDS1): a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Haematol. 2024;11:e38–50. doi: 10.1016/S2352-3026(23)00333-2. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
