High frequency CCR5 editing in human hematopoietic stem progenitor cells protects xenograft mice from HIV infection
- PMID: 39774003
- PMCID: PMC11707138
- DOI: 10.1038/s41467-025-55873-3
High frequency CCR5 editing in human hematopoietic stem progenitor cells protects xenograft mice from HIV infection
Abstract
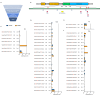
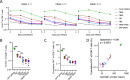
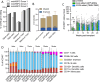
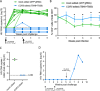
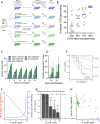
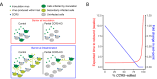
The only cure of HIV has been achieved in a small number of people who received a hematopoietic stem cell transplant (HSCT) comprising allogeneic cells carrying a rare, naturally occurring, homozygous deletion in the CCR5 gene. The rarity of the mutation and the significant morbidity and mortality of such allogeneic transplants precludes widespread adoption of this HIV cure. Here, we show the application of CRISPR/Cas9 to achieve >90% CCR5 editing in human, mobilized hematopoietic stem progenitor cells (HSPC), resulting in a transplant that undergoes normal hematopoiesis, produces CCR5 null T cells, and renders xenograft mice refractory to HIV infection. Titration studies transplanting decreasing frequencies of CCR5 edited HSPCs demonstrate that <90% CCR5 editing confers decreasing protective benefit that becomes negligible between 54% and 26%. Our study demonstrates the feasibility of using CRISPR/Cas9/RNP to produce an HSPC transplant with high frequency CCR5 editing that is refractory to HIV replication. These results raise the potential of using CRISPR/Cas9 to produce a curative autologous HSCT and bring us closer to the development of a cure for HIV infection.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: ZD is a current employee, holds stock and stock options in CRISPR Therapeutics AG. TB and TH are former employees and shareholders of CRISPR Therapeutics AG. The other authors declare no competing interests.
Figures


Similar articles
-
CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance In Vivo.Mol Ther. 2017 Aug 2;25(8):1782-1789. doi: 10.1016/j.ymthe.2017.04.027. Epub 2017 May 17. Mol Ther. 2017. PMID: 28527722 Free PMC article.
-
A simultaneous knockout knockin genome editing strategy in HSPCs potently inhibits CCR5- and CXCR4-tropic HIV-1 infection.Cell Stem Cell. 2024 Apr 4;31(4):499-518.e6. doi: 10.1016/j.stem.2024.03.002. Cell Stem Cell. 2024. PMID: 38579682 Free PMC article.
-
Multilayered HIV-1 resistance in HSPCs through CCR5 Knockout and B cell secretion of HIV-inhibiting antibodies.Nat Commun. 2025 Apr 1;16(1):3103. doi: 10.1038/s41467-025-58371-8. Nat Commun. 2025. PMID: 40164595 Free PMC article.
-
Killing two birds with one stone: CRISPR/Cas9 CCR5 knockout hematopoietic stem cells transplantation to treat patients with HIV infection and hematological malignancies concurrently.Clin Exp Med. 2023 Dec;23(8):4163-4175. doi: 10.1007/s10238-023-01129-7. Epub 2023 Jul 27. Clin Exp Med. 2023. PMID: 37500934 Review.
-
The Strategies and Challenges of CCR5 Gene Editing in Hematopoietic Stem and Progenitor Cells for the Treatment of HIV.Stem Cell Rev Rep. 2021 Oct;17(5):1607-1618. doi: 10.1007/s12015-021-10145-7. Epub 2021 Mar 31. Stem Cell Rev Rep. 2021. PMID: 33788143 Review.
Cited by
-
CRISPR/Cas9 for achieving postintervention HIV control.Curr Opin HIV AIDS. 2025 Sep 1;20(5):432-440. doi: 10.1097/COH.0000000000000963. Epub 2025 Jul 18. Curr Opin HIV AIDS. 2025. PMID: 40682401 Free PMC article. Review.
-
CCR5 gene editing and HIV immunotherapy: current understandings, challenges, and future directions.Front Immunol. 2025 Jun 18;16:1590690. doi: 10.3389/fimmu.2025.1590690. eCollection 2025. Front Immunol. 2025. PMID: 40607431 Free PMC article.
References
-
- Siliciano, J. D. et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med.9, 727–728 (2003). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

