Activity of the mammalian DNA transposon piggyBat from Myotis lucifugus is restricted by its own transposon ends
- PMID: 39774116
- PMCID: PMC11707139
- DOI: 10.1038/s41467-024-55784-9
Activity of the mammalian DNA transposon piggyBat from Myotis lucifugus is restricted by its own transposon ends
Abstract
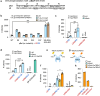
Members of the piggyBac superfamily of DNA transposons are widely distributed in host genomes ranging from insects to mammals. The human genome has retained five piggyBac-derived genes as domesticated elements although they are no longer mobile. Here, we have investigated the transposition properties of piggyBat from Myotis lucifugus, the only known active mammalian DNA transposon, and show that its low activity in human cells is due to subterminal inhibitory DNA sequences. Activity can be dramatically improved by their removal, suggesting the existence of a mechanism for the suppression of transposon activity. The cryo-electron microscopy structure of the piggyBat transposase pre-synaptic complex showed an unexpected mode of DNA binding and recognition using C-terminal domains that are topologically different from those of the piggyBac transposase. Here we show that structure-based rational re-engineering of the transposase through the removal of putative phosphorylation sites and a changed domain organization - in combination with truncated transposon ends - results in a transposition system that is at least 100-fold more active than wild-type piggyBat.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: This work was partially funded by a Collaborative Research and Development Agreement DK22-1043 between the NIDDK and SalioGen Therapeutics. M.H.W. previously served on the scientific advisory board of SalioGen. A.B.H., C.M.F., and F.D. have submitted a patent application (No. 63/632,275) that covers modifications to the piggyBat transposon system that increase its activity. The remaining authors declare no competing interests.
Figures







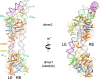
References
-
- Craig, N. L. in Mobile DNA III, ed. Craig, Chandler, Gellert, Lambowitz, Rice, & Sandmeyer, 3–39 (American Society of Microbiology, 2015).
-
- Ozata, D. M., Gainetdinov, I., Zoch, A., O’Carroll, D. & Zamore, P. D. PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet.20, 89–108 (2019). - PubMed
-
- Ivics, Z., Hackett, P. B., Plasterk, R. H. & Izsvák, Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell91, 501–510 (1997). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

