Phase separation of initiation hubs on cargo is a trigger switch for selective autophagy
- PMID: 39774270
- PMCID: PMC11821514
- DOI: 10.1038/s41556-024-01572-y
Phase separation of initiation hubs on cargo is a trigger switch for selective autophagy
Abstract
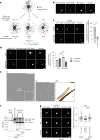
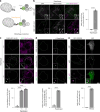
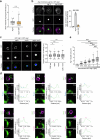
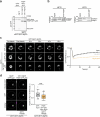
Autophagy is a key cellular quality control mechanism. Nutrient stress triggers bulk autophagy, which nonselectively degrades cytoplasmic material upon formation and liquid-liquid phase separation of the autophagy-related gene 1 (Atg1) complex. In contrast, selective autophagy eliminates protein aggregates, damaged organelles and other cargoes that are targeted by an autophagy receptor. Phase separation of cargo has been observed, but its regulation and impact on selective autophagy are poorly understood. Here, we find that key autophagy biogenesis factors phase separate into initiation hubs at cargo surfaces in yeast, subsequently maturing into sites that drive phagophore nucleation. This phase separation is dependent on multivalent, low-affinity interactions between autophagy receptors and cargo, creating a dynamic cargo surface. Notably, high-affinity interactions between autophagy receptors and cargo complexes block initiation hub formation and autophagy progression. Using these principles, we converted the mammalian reovirus nonstructural protein µNS, which accumulates as particles in the yeast cytoplasm that are not degraded, into a neo-cargo that is degraded by selective autophagy. We show that initiation hubs also form on the surface of different cargoes in human cells and are key to establish the connection to the endoplasmic reticulum, where the phagophore assembly site is formed to initiate phagophore biogenesis. Overall, our findings suggest that regulated phase separation underscores the initiation of both bulk and selective autophagy in evolutionarily diverse organisms.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures












References
-
- Fujioka, Y. et al. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat. Struct. Mol. Biol.21, 513–521 (2014). - PubMed
MeSH terms
Substances
Grants and funding
- 259130777/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 423813989/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 403222702/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 390939984/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450216812/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 409673687/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 769065/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 101041982/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

