tomoseqr: A Bioconductor package for spatial reconstruction and visualization of 3D gene expression patterns based on RNA tomography
- PMID: 39774435
- PMCID: PMC11709299
- DOI: 10.1371/journal.pone.0311296
tomoseqr: A Bioconductor package for spatial reconstruction and visualization of 3D gene expression patterns based on RNA tomography
Abstract
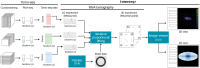
RNA tomography computationally reconstructs 3D spatial gene expression patterns genome-widely from 1D tomo-seq data, generated by RNA sequencing of cryosection samples along three orthogonal axes. We developed tomoseqr, an R package designed for RNA tomography analysis of tomo-seq data, to reconstruct and visualize 3D gene expression patterns through user-friendly graphical interfaces. We show the effectiveness of tomoseqr using simulated and real tomo-seq data, validating its utility for researchers. R package tomoseqr is available on Bioconductor (https://doi.org/doi:10.18129/B9.bioc.tomoseqr) and GitHub (https://github.com/bioinfo-tsukuba/tomoseqr).
Copyright: © 2025 Matsuzawa et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

