Prediction model establishment of prognosis factors for acute myeloid leukemia based on the SEER database
- PMID: 39774789
- PMCID: PMC11707327
- DOI: 10.1038/s41598-025-85310-w
Prediction model establishment of prognosis factors for acute myeloid leukemia based on the SEER database
Abstract
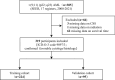
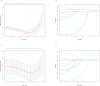
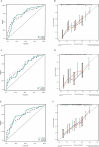
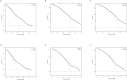
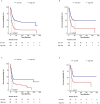
Acute myeloid leukemia (AML) with t (9;11) (p22; q23) presents as a varied hematological malignancy. The t (9;11) (p22; q23) translocation is the most common among 11q23/KMT2A rearrangements in AML. This research aimed to develop a nomogram for precise prediction of overall survival (OS) and cancer-specific survival (CSS) in AML with the t (9;11) (p22; q23) translocation. We utilized the Surveillance, Epidemiology, and End Results (SEER) database to identify patients diagnosed with t (9;11) (p22; q23) AML from 2000 to 2021. Prognostic factors for this AML subtype were determined using least absolute shrinkage and selection operator (LASSO) regression, which guided the creation of prognostic nomograms. To evaluate the model's discrimination, accuracy, and effectiveness, we employed the concordance index (C-index), calibration charts, receiver operating characteristic curves (ROC), area under the curve (AUC), and decision-curve analysis (DCA). The research was meticulously planned, executed, and documented in full adherence to the TRIPOD guidelines. The nomogram was developed using key variables including age, race, first primary tumor, and chemotherapy. The concordance indices (C-indices) were 0.704 for OS and for 0.686 for CSS. Patients were classified into high-risk and low-risk groups based on nomogram scores, with significant differences in OS and CSS between these groups (P < 0.001). This study developed innovative nomograms that combine clinical and treatment factors to predict 1-, 3-, and 5-year survival rates for patients with t (9;11) (p22; q23) AML.
Keywords: Analysis; Nomogram; Prognosis; SEER; T (911) (p22q23) acute myeloid leukemia.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The data analyzed and used in this study was obtained from Surveillance, Epidemiology, and End Results (SEER) database in accordance with the SEER data use agreement. Therefore, this study did not require approval of ethical board. Competing interests: The authors declare no competing interests. Consent for publication: Not applicable.
Figures



References
-
- Tirado, C. A. et al. A t(9;11)(p22;q23) within the context of a complex karyotype is associated with a poor prognosis in a 19-year-old patient with AML. J. Assoc. Genet. Technol.47, 78–83 (2021). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

