Spatially Resolved Tumor Ecosystems and Cell States in Gastric Adenocarcinoma Progression and Evolution
- PMID: 39774838
- PMCID: PMC11962405
- DOI: 10.1158/2159-8290.CD-24-0605
Spatially Resolved Tumor Ecosystems and Cell States in Gastric Adenocarcinoma Progression and Evolution
Abstract
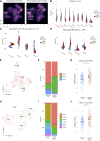
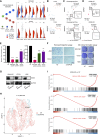
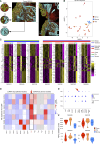
Integration of spatial transcriptomic (GeoMx Digital Spatial Profiler) and single-cell RNA sequencing data from multiple gastric cancers identifies spatially resolved expression-based intratumoral heterogeneity, associated with distinct immune microenvironments. We uncovered two separate evolutionary trajectories associated with specific molecular subtypes, clinical prognoses, stromal neighborhoods, and genetic drivers. Tumor-stroma interfaces emerged as a unique state of tumor ecology.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
J.J. Zhao reports grants from the National University Health System Seed Fund (NUHSRO/2024/008/RO5+6/Seed-Sep23/01), National University Hospital Junior Research Award 2023 (JRA/Sep23/002), Chan Heng Leong Education & Research Fund 2024 award by the National University Hospital Singapore, 2025 Conquer Cancer Merit Award by Conquer Cancer, the ASCO Foundation, and Dean’s Research Development Award awarded by the Yong Loo Lin School of Medicine, National University of Singapore, outside the submitted work. R. Sundar reports grants from the National Medical Research Council during the conduct of the study and other support from Astellas, AstraZeneca, Bayer, BeiGene, Bristol Myers Squibb, Daiichi Sankyo, DKSH, Eisai, GSK, Merck, MSD, Novartis, Pierre-Fabre, Sanofi, Taiho, Tavotek, Eli Lilly, Ipsen, Roche, CytoMed, Paxman, and Teladoc outside the submitted work. No disclosures were reported by the other authors.
Figures




References
-
- Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, et al. Cancer statistics for the year 2020: an overview. Int J Cancer 2021;149:778–89. - PubMed
-
- Alsina M, Arrazubi V, Diez M, Tabernero J. Current developments in gastric cancer: from molecular profiling to treatment strategy. Nat Rev Gastroenterol Hepatol 2023;20:155–70. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

