Asciminib resistance of a new BCR::ABL1 p.I293_K294insSSLRD mutant detected in a Ph + ALL patient
- PMID: 39774950
- PMCID: PMC11971149
- DOI: 10.1007/s00277-024-06142-8
Asciminib resistance of a new BCR::ABL1 p.I293_K294insSSLRD mutant detected in a Ph + ALL patient
Abstract
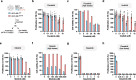
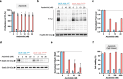
Chronic myeloid leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia patients largely benefit from an expanding tyrosine kinase inhibitors (TKIs) toolbox that has improved the outcome of both diseases. However, TKI success is continuously challenged by mutation-driven acquired resistance and therefore, close monitoring of clonal genetic diversity is necessary to ensure proper clinical management and adequate response to treatment. Here, we report the case of a ponatinib-resistant Philadelphia chromosome-positive acute lymphoblastic leukemia (Ph + ALL) patient harboring a BCR::ABL1 p.I293_K294insSLLRD mutation. Using in vitro proliferation assays on newly generated Ba/F3 cell lines, we confirmed that the mutation confers moderate resistance to ponatinib, and to imatinib and nilotinib. In contrast, BCR::ABL1SLLRD Ba/F3 cells remain highly sensitive to dasatinib. Unexpectedly, the insertion also provides resistance to asciminib with no inhibitory effect up to 1000 nM. Based on predicted structural models, we speculate that the p.I293_K294insSLLRD disrupts the interaction between the SH3 domain and the kinase domain, shifting the equilibrium toward the active conformation. This shift confers resistance to TKIs that preferentially bind to the inactive conformation, as well as to the allosteric asciminib inhibitor. However, the mutation retains sensitivity to dasatinib, which targets the active form of the kinase.
Keywords: ALL; BCR::ABL1; CML; Cancer resistance; Insertion mutation; Tyrosine kinase inhibitor.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Compliance with ethical standards: The patient involved in the case report described in this study signed an informed consent as part of the GRAAPH protocol (ClinicalTrials.gov Identifier: NCT00327678). Competing interests: S.D. has received honorarium from Novartis and Incyte, for speaking engagements. All other authors declare no conflict of interests.
Figures

References
-
- Faderl S, Talpaz M, Estrov Z et al (1999) The Biology of Chronic Myeloid Leukemia. N Engl J Med 341(3):164–172. 10.1056/NEJM199907153410306 - PubMed
-
- Höglund M, Sandin F, Simonsson B (2015) Epidemiology of chronic myeloid leukaemia: an update. Ann Hematol 94(Suppl 2):S241-247. 10.1007/s00277-015-2314-2 - PubMed
-
- Secker-Walker LM, Craig JM, Hawkins JM et al (1991) Philadelphia positive acute lymphoblastic leukemia in adults: age distribution. BCR breakpoint and prognostic significance Leukemia 5(3):196–199 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

