Multi-omics-based mapping of decidualization resistance in patients with a history of severe preeclampsia
- PMID: 39775038
- PMCID: PMC11835751
- DOI: 10.1038/s41591-024-03407-7
Multi-omics-based mapping of decidualization resistance in patients with a history of severe preeclampsia
Abstract
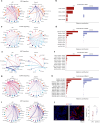
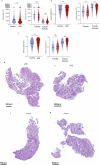
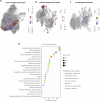
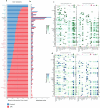
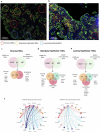
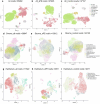
Endometrial decidualization resistance (DR) is implicated in various gynecological and obstetric conditions. Here, using a multi-omic strategy, we unraveled the cellular and molecular characteristics of DR in patients who have suffered severe preeclampsia (sPE). Morphological analysis unveiled significant glandular anatomical abnormalities, confirmed histologically and quantified by the digitization of hematoxylin and eosin-stained tissue sections. Single-cell RNA sequencing (scRNA-seq) of endometrial samples from patients with sPE (n = 11) and controls (n = 12) revealed sPE-associated shifts in cell composition, manifesting as a stromal mosaic state characterized by proliferative stromal cells (MMP11 and SFRP4) alongside IGFBP1+ decidualized cells, with concurrent epithelial mosaicism and a dearth of epithelial-stromal transition associated with decidualization. Cell-cell communication network mapping underscored aberrant crosstalk among specific cell types, implicating crucial pathways such as endoglin, WNT and SPP1. Spatial transcriptomics in a replication cohort validated DR-associated features. Laser capture microdissection/mass spectrometry in a second replication cohort corroborated several scRNA-seq findings, notably the absence of stromal to epithelial transition at a pathway level, indicating a disrupted response to steroid hormones, particularly estrogens. These insights shed light on potential molecular mechanisms underpinning DR pathogenesis in the context of sPE.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures










References
-
- Moreno, I. et al. The human periconceptional maternal-embryonic space in health and disease. Physiol. Rev.103, 1965–2038 (2023). - PubMed
-
- Wang, W. et al. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat. Med.26, 1644–1653 (2020). - PubMed
-
- Tabanelli, S., Tang, B. & Gurpide, E. In vitro decidualization of human endometrial stromal cells. J. Steroid Biochem. Mol. Biol.42, 337–344 (1992). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

