Longitudinal monitoring of sewershed resistomes in socioeconomically diverse urban neighborhoods
- PMID: 39775111
- PMCID: PMC11707339
- DOI: 10.1038/s43856-024-00729-y
Longitudinal monitoring of sewershed resistomes in socioeconomically diverse urban neighborhoods
Abstract
Background: Understanding factors associated with antimicrobial resistance (AMR) distribution across populations is a necessary step in planning mitigation measures. While associations between AMR and socioeconomic-status (SES), including employment and education have been increasingly recognized in low- and middle-income settings, connections are less clear in high-income countries where SES remains an important influence on other health outcomes.
Methods: We explored the relationship between SES and AMR in Calgary, Canada using spatially-resolved wastewater-based surveillance of resistomes detected by metagenomics across eight socio-economically diverse urban neighborhoods. Resistomes were established by shotgun-sequencing of wastewater pellets, and qPCR of targeted-AMR genes. SES status was established using 2021 Canadian census data. Conducting this comparison during the height of COVID-related international travel restrictions (Dec. 2020-Oct. 2021) allowed the hypotheses linking SES and AMR to be assessed with limited confounding. These were compared with sewage metagenomes from 244 cities around the world, linked with Human Development Index (HDI).
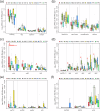
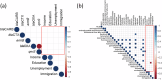
Results: Wastewater metagenomes from Calgary's socioeconomically diverse neighborhoods exhibit highly similar resistomes, with no quantitative differences (p > 0.05), low Bray-Curtis dissimilarity, and no significant correlations with SES. By comparison, dissimilarity is observed between globally-sourced resistomes (p < 0.05), underscoring the homogeneity of resistomes in Calgary's sub-populations. The analysis of globally-sourced resistomes alongside Calgary's resistome further reveals lower AMR burden in Calgary relative to other cities around the world. This is particularly pronounced for the most clinically-relevant AMR genes (e.g., beta-lactamases, macrolide-lincosamide-streptogramin).
Conclusions: This work showcases the effectiveness of inclusive and comprehensive wastewater-based surveillance for exploring the interplay between SES and AMR.
Plain language summary
Antimicrobial resistance (AMR) occurs when antimicrobial treatments fail to work and microbes continue to grow. This is a result of microbes acquiring AMR genes. Antimicrobial resistance (AMR) is an increasing public health threat. Some studies have suggested an association between AMR and socioeconomic factors. The amount of AMR can be monitored by investigating the presence of specific genes indicative of AMR in wastewater. To explore this within a high-income country with publicly funded health care, we collected wastewater from eight socioeconomically diverse neighborhoods across a large Canadian city. Conducted over eleven months during COVID-19-related travel restrictions, we did not observe an association between socioeconomic status of residents and the amount or types of AMR genes in wastewater. We also compared AMR genes from wastewater from cities across the globe, where we observed the presence of AMR genes significantly differed along established socio-economic parameters. Overall, our findings revealed the relationship between AMR genes and socioeconomic factors is dynamic, and context dependent.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Collignon, P., Beggs, J. J., Walsh, T. R., Gandra, S. & Laxminarayan, R. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. Lancet Planet. Heath2, e398–e405 (2018). - PubMed
-
- Wozniak, T. M., Cuningham, W., Ledingham, K. & McCulloch, K. Contribution of socio-economic factors in the spread of antimicrobial resistant infections in Australian primary healthcare clinics. J. Global Antimicrob. Resist.30, 294–301 (2022). - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

