Plasmodium falciparum surf4.1 in clinical isolates: From genetic variation and variant diversity to in silico design immunopeptides for vaccine development
- PMID: 39775228
- PMCID: PMC11684625
- DOI: 10.1371/journal.pone.0312091
Plasmodium falciparum surf4.1 in clinical isolates: From genetic variation and variant diversity to in silico design immunopeptides for vaccine development
Abstract
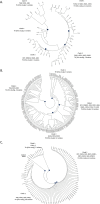
SURFINs protein family expressed on surface of both infected red blood cell and merozoite surface making them as interesting vaccine candidate for erythrocytic stage of malaria infection. In this study, we analyze genetic variation of Pfsurf4.1 gene, copy number variation, and frequency of SURFIN4.1 variants of P. falciparum in clinical isolates. In addition, secondary structure prediction and immunoinformatic were employed to identify immunogenic epitopes in humoral response as proposed vaccine candidates. Overall, our data demonstrate extensive polymorphism of SURFIN4.1 in both genetic and protein level. The surf4.1 gene showed extensive genetic variation with total of 447 polymorphic sites with maximum of three variants as well as singlet/triplet bases indels and mini/microsatellites in the coding sequence. The exon1 encoding extracellular region exhibited higher variation compared to exon2 which coding for intracellular domain. Interestingly, selective pressure was detected on both extracellular region (Var1 and Var2) as well as intracellular region (WRD2 and WRD3). Importantly, extensive full gene analysis suggests adenosine insertion at three key points nucleotide bases (nt 2409/2410, 3809/3810, and 4439/4440) of exon2 could lead to frameshift mutation resulted in four different SURFIN4.1 variants (TMs, WD1, WD2 and WD3). The SURFIN4.1 variant TMs was the most observed type with 67% frequency (51/76). Along with more than one copy number of surf4.1 gene was observed with frequency of 13% (9/70). Despite substantial polymorphism, analysis of relatedness within P. falciparum population using full coding sequence was able to group SURFIN4.1 protein into five distinct clades and reduced into four clades when using only exon1 coding sequence. Also, predicted secondary structure revealed conserved structure of five helix domains of extracellular region which similar among four SURFIN4.1 variant types. In addition, in silico design eight immunopeptides derived from SURFIN4.1, four of which are highly conserved and four of dimorphic epitopes, as potential vaccine candidates.
Copyright: © 2024 Noranate et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Nilsson Bark SK, Ahmad R, Dantzler K, Lukens AK, De Niz M, Szucs MJ, et al. Quantitative Proteomic Profiling Reveals Novel Plasmodium falciparum Surface Antigens and Possible Vaccine Candidates. Mol Cell Proteomics. 2018;17(1):43–60. Epub 2017/11/23. doi: 10.1074/mcp.RA117.000076 ; PubMed Central PMCID: PMC5750850. - DOI - PMC - PubMed
-
- Winter G, Kawai S, Haeggstrom M, Kaneko O, von Euler A, Kawazu S, et al. SURFIN is a polymorphic antigen expressed on Plasmodium falciparum merozoites and infected erythrocytes. J Exp Med. 2005;201(11):1853–63. Epub 2005/06/09. doi: 10.1084/jem.20041392 ; PubMed Central PMCID: PMC2213267. - DOI - PMC - PubMed
-
- Quintana MDP, Ch’ng JH, Moll K, Zandian A, Nilsson P, Idris ZM, et al. Antibodies in children with malaria to PfEMP1, RIFIN and SURFIN expressed at the Plasmodium falciparum parasitized red blood cell surface. Sci Rep. 2018;8(1):3262. Epub 2018/02/21. doi: 10.1038/s41598-018-21026-4 ; PubMed Central PMCID: PMC5818650. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

