Withaferin A inhibits Chikungunya virus nsP2 protease and shows antiviral activity in the cell culture and mouse model of virus infection
- PMID: 39775571
- PMCID: PMC11723598
- DOI: 10.1371/journal.ppat.1012816
Withaferin A inhibits Chikungunya virus nsP2 protease and shows antiviral activity in the cell culture and mouse model of virus infection
Abstract
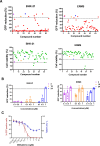
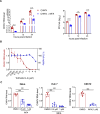
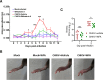
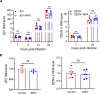
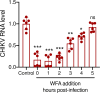
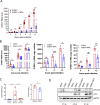
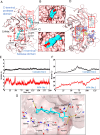
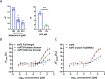
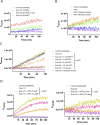
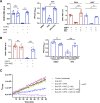
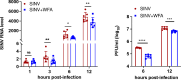
Chikungunya virus (CHIKV) is a mosquito-transmitted alphavirus causing fever, myalgia, and debilitating joint swelling and pain, which in many patients becomes chronic. The frequent epidemics of CHIKV across the world pose a significant public health burden necessitating the development of effective antiviral therapeutics. A cellular imaging-based high-content screening of natural compounds identified withaferin A (WFA), a steroidal lactone isolated from the plant Withania somnifera, as a potent antiviral against CHIKV. In the ERMS cells, WFA inhibited CHIKV replication early during the life cycle by binding the CHIKV non-structural protein nsP2 and inhibiting its protease activity. This inhibited the viral polyprotein processing and the minus-sense viral RNA synthesis. WFA mounted the nsP2 protease inhibitory activity through its oxidising property as the reducing agents N-acetylcysteine and Glutathione-monoethyl ester effectively reversed the WFA-mediated protease inhibition in vitro and abolished the WFA-mediated antiviral activity in cultured cells. WFA inhibited CHIKV replication in the C57BL/6 mouse model of chikungunya disease, resulting in significantly lower viremia. Importantly, CHIKV-infected mice showed significant joint swelling which was not seen in WFA-treated mice. These data demonstrate the potential of WFA as a novel CHIKV antiviral.
Copyright: © 2024 Sharma et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- ECDC. 2023. European Centre for Disease Prevention and Control. https://www.ecdc.europa.eu/. Accessed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

