Evidence for gene essentiality in Leishmania using CRISPR
- PMID: 39775585
- PMCID: PMC11684651
- DOI: 10.1371/journal.pone.0316331
Evidence for gene essentiality in Leishmania using CRISPR
Abstract
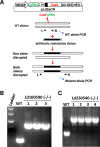
The ability to determine the essentiality of a gene in the protozoan parasite Leishmania is important to identify potential targets for intervention and understanding the parasite biology. CRISPR gene editing technology has significantly improved gene targeting efficiency in Leishmania. There are two commonly used CRISPR gene targeting methods in Leishmania; the stable expression of the gRNA and Cas9 using a plasmid containing a Leishmania ribosomal RNA gene promoter (rRNA-P stable protocol) and the T7 RNA polymerase based transient gRNA expression system in promastigotes stably expressing Cas9 (T7 transient protocol). There are distinct advantages with both systems. The T7 transient protocol is excellent for high throughput gene deletions and has been used to successfully delete hundreds of Leishmania genes to study mutant phenotypes and several research labs are now using this protocol to target all the genes in L. mexicana genome. The rRNA-P stable protocol stably expresses the plasmid derived gRNA and has been used to delete or disrupt single and multicopy Leishmania genes, perform single nucleotide changes and provide evidence for gene essentiality by directly observing null mutant promastigotes dying in culture. In this study, the rRNA-P stable protocol was used to target 22 Leishmania genes in which null mutants were not generated using the T7 transient protocol. Notably, the rRNA-P stable protocol was able to generate alive null mutants for 8 of the 22 genes. These results demonstrate the rRNA-P stable protocol could be used alone or in combination with the T7 transient protocol to investigate gene essentiality in Leishmania.
Copyright: © 2024 Zhang, Matlashewski. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Application of CRISPR/Cas9-Mediated Genome Editing in Leishmania.Methods Mol Biol. 2020;2116:199-224. doi: 10.1007/978-1-0716-0294-2_14. Methods Mol Biol. 2020. PMID: 32221923
-
gRNA-transient expression system for simplified gRNA delivery in CRISPR/Cas9 genome editing.J Biosci Bioeng. 2019 Sep;128(3):373-378. doi: 10.1016/j.jbiosc.2019.02.009. Epub 2019 Apr 19. J Biosci Bioeng. 2019. PMID: 31010727
-
CRISPR-Cas9-Mediated Genome Editing in Leishmania donovani.mBio. 2015 Jul 21;6(4):e00861. doi: 10.1128/mBio.00861-15. mBio. 2015. PMID: 26199327 Free PMC article.
-
Optimization of genome editing through CRISPR-Cas9 engineering.Bioengineered. 2016 Apr;7(3):166-74. doi: 10.1080/21655979.2016.1189039. Bioengineered. 2016. PMID: 27340770 Free PMC article. Review.
-
CRISPR/Cas9 Guide RNA Design Rules for Predicting Activity.Methods Mol Biol. 2020;2115:351-364. doi: 10.1007/978-1-0716-0290-4_19. Methods Mol Biol. 2020. PMID: 32006410 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

