Spatiotemporal dynamics of fetal liver hematopoietic niches
- PMID: 39775824
- PMCID: PMC11706214
- DOI: 10.1084/jem.20240592
Spatiotemporal dynamics of fetal liver hematopoietic niches
Abstract
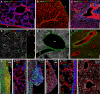
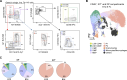
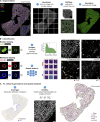
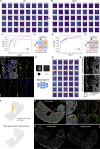
Embryonic hematopoietic cells develop in the fetal liver (FL), surrounded by diverse non-hematopoietic stromal cells. However, the spatial organization and cytokine production patterns of the stroma during FL development remain poorly understood. Here, we characterized and mapped the hematopoietic and stromal cell populations at early (E12.5-14.5) FL stages, revealing that while hepatoblasts were the primary source of hematopoietic growth factors, other stromal cells-including mesenchymal, mesothelial, and endothelial cells-also contributed to this signaling network. Using a dedicated image analysis pipeline, we quantified cell distances to tissue structures and defined neighbor relationships, uncovering that different hematopoietic progenitors exhibit distinct preferences for neighboring stromal cells and show developmental changes in spatial distribution. Notably, our data suggest that the sub-mesothelium region plays a prominent role in early fetal hematopoiesis. This approach offers a valuable tool for studying complex cellular interactions in biological systems, providing new insights into hematopoietic niche organization during development.
© 2025 Mesquita Peixoto et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures





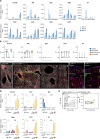

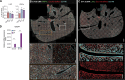


References
-
- Breiteneder-Geleff, S., Soleiman A., Kowalski H., Horvat R., Amann G., Kriehuber E., Diem K., Weninger W., Tschachler E., Alitalo K., and Kerjaschki D.. 1999. Angiosarcomas express mixed endothelial phenotypes of blood and lymphatic capillaries: Podoplanin as a specific marker for lymphatic endothelium. Am. J. Pathol. 154:385–394. 10.1016/S0002-9440(10)65285-6 - DOI - PMC - PubMed
-
- Buono, M., Facchini R., Matsuoka S., Thongjuea S., Waithe D., Luis T.C., Giustacchini A., Besmer P., Mead A.J., Jacobsen S.E.W., and Nerlov C.. 2016. A dynamic niche provides kit ligand in a stage-specific manner to the earliest thymocyte progenitors. Nat. Cell Biol. 18:157–167. 10.1038/ncb3299 - DOI - PMC - PubMed
-
- Chea, S., Schmutz S., Berthault C., Perchet T., Petit M., Burlen-Defranoux O., Goldrath A.W., Rodewald H.-R., Cumano A., and Golub R.. 2016. Single-cell gene expression analyses reveal heterogeneous responsiveness of fetal innate lymphoid progenitors to notch signaling. Cell Rep. 14:1500–1516. 10.1016/j.celrep.2016.01.015 - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

