Direct Measurement of the Mutation Rate and Its Evolutionary Consequences in a Critically Endangered Mollusk
- PMID: 39775835
- PMCID: PMC11704959
- DOI: 10.1093/molbev/msae266
Direct Measurement of the Mutation Rate and Its Evolutionary Consequences in a Critically Endangered Mollusk
Abstract
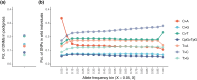
The rate at which mutations arise is a fundamental parameter of biology. Despite progress in measuring germline mutation rates across diverse taxa, such estimates are missing for much of Earth's biodiversity. Here, we present the first estimate of a germline mutation rate from the phylum Mollusca. We sequenced three pedigreed families of the white abalone Haliotis sorenseni, a long-lived, large-bodied, and critically endangered mollusk, and estimated a de novo mutation rate of 8.60 × 10-9 single nucleotide mutations per site per generation. This mutation rate is similar to rates measured in vertebrates with comparable generation times and longevity to abalone, and higher than mutation rates measured in faster-reproducing invertebrates. The spectrum of de novo mutations is also similar to that seen in vertebrate species, although an excess of rare C > A polymorphisms in wild individuals suggests that a modifier allele or environmental exposure may have once increased C > A mutation rates. We use our rate to infer baseline effective population sizes (Ne) across multiple Pacific abalone and find that abalone persisted over most of their evolutionary history as large and stable populations, in contrast to extreme fluctuations over recent history and small census sizes in the present day. We then use our mutation rate to infer the timing and pattern of evolution of the abalone genus Haliotis, which was previously unknown due to few fossil calibrations. Our findings are an important step toward understanding mutation rate evolution and they establish a key parameter for conservation and evolutionary genomics research in mollusks.
Keywords: conservation; mollusk; mutation rate; phylogenomics.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures



References
-
- Andrews AH, Leaf RT, Rogers-Bennett L, Neuman M, Hawk H, Cailliet GM. Bomb radiocarbon dating of the endangered white abalone (Haliotis sorenseni): investigations of age, growth and lifespan. Mar Freshw Res. 2013:64(11):1029–1039. 10.1071/MF13007. - DOI
-
- Babcock R, Keesing J. Fertilization biology of the abalone Haliotis laevigata: laboratory and field studies. Can J Fish Aquat Sci. 1999:56(9):1668–1678. 10.1139/f99-106. - DOI
-
- Bánki O, Roskov Y, Döring M, Ower G, Hernández Robles DR, Plata Corredor CA, Stjernegaard Jeppesen T, Örn A, Vandepitte L, Hobern D, et al. Catalogue of life (version 2024-07-18). Amsterdam (NL): Catalogue of Life; 2024.

