Development and validation of a molecular classifier of meningiomas
- PMID: 39775867
- PMCID: PMC12187452
- DOI: 10.1093/neuonc/noae242
Development and validation of a molecular classifier of meningiomas
Abstract
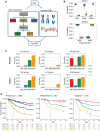
Background: Meningiomas exhibit considerable clinical and biological heterogeneity. We previously identified 4 distinct molecular groups (immunogenic, NF2-wildtype, hypermetabolic, and proliferative) that address much of this heterogeneity. Despite the utility of these groups, the stochasticity of clustering methods and the use of multi-omics data for discovery limits the potential for classifying prospective cases. We sought to address this with a dedicated classifier.
Methods: Using an international cohort of 1698 meningiomas, we constructed and rigorously validated a machine learning-based molecular classifier using only DNA methylation data as input. Original and newly predicted molecular groups were compared using DNA methylation, RNA sequencing, copy number profiles, whole-exome sequencing, and clinical outcomes.
Results: We show that group-specific outcomes in the validation cohort are nearly identical to those originally described, with median progression-free survival (PFS) of 7.4 (4.9-Inf) years in hypermetabolic tumors and 2.5 (2.3-5.3) years in proliferative tumors (not reached in the other groups). Tumors classified as NF2-wildtype had no NF2 mutations, and 51.4% had canonical mutations previously described in this group. RNA pathway analysis revealed upregulation of immune-related pathways in the immunogenic group, metabolic pathways in the hypermetabolic group, and cell cycle programs in the proliferative group. Bulk deconvolution similarly revealed the enrichment of macrophages in immunogenic tumors and neoplastic cells in hypermetabolic and proliferative tumors with similar proportions to those originally described.
Conclusions: Our DNA methylation-based classifier, which is publicly available for immediate clinical use, recapitulates the biology and outcomes of the original molecular groups as assessed using multiple metrics/platforms that were not used in its training.
Keywords: DNA methylation; meningioma | molecular classification | Neuro-Oncology | outcome prediction.
© The Author(s) 2025. Published by Oxford University Press on behalf of the Society for Neuro-Oncology.
Conflict of interest statement
The authors of this manuscript have no conflicts of interest to disclose.
Figures

References
-
- Sahm F, Schrimpf D, Stichel D, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18(5):682–694. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA263196/CA/NCI NIH HHS/United States
- Brain Tumour Charity
- HHSN261201500003I/NH/NIH HHS/United States
- HHSN26100039/NH/NIH HHS/United States
- RN482811-481519/CAPMC/ CIHR/Canada
- GN-00043/Brain Tumor Charity United Kingdom
- GN-000693/Brain Tumor Charity United Kingdom
- the UHN Foundation, Mary Hunter Meningioma Research Fund
- V Foundation for Cancer Research
- the CIHR Vanier Scholarship
- the American Association of Neurological Surgeons Neurosurgery Education & Research Foundation Research Fellowship
- the Congress of Neurological Surgeons Tumor Section
- the Princess Margaret Hospital Foundation Hold 'em For Life Oncology Fellowship
LinkOut - more resources
Full Text Sources
Miscellaneous

