Machine Learning of Endoscopy Images to Identify, Classify, and Segment Sinonasal Masses
- PMID: 39776302
- PMCID: PMC12048764
- DOI: 10.1002/alr.23525
Machine Learning of Endoscopy Images to Identify, Classify, and Segment Sinonasal Masses
Abstract
Background: We developed and assessed the performance of a machine learning model (MLM) to identify, classify, and segment sinonasal masses based on endoscopic appearance.
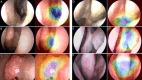
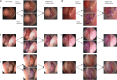
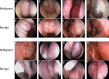
Methods: A convolutional neural network-based model was constructed from nasal endoscopy images from patients evaluated at an otolaryngology center between 2013 and 2024. Images were classified into four groups: normal endoscopy, nasal polyps, benign, and malignant tumors. Polyps and tumors were confirmed with histopathological diagnosis. Images were annotated by an otolaryngologist and independently verified by two other otolaryngologists. We used high- and low-quality images to mirror real-world conditions. The models used for classification (EfficientNet-B2) and segmentation (nnUNet) were trained, validated, and tested at an 8:1:1 ratio. The performance accuracy was averaged across a 10-fold cross-validation assessment. Segmentation accuracy was assessed via Dice similarity coefficients.
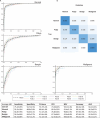
Results: A total of 1242 images from 311 patients were used. The MLM was trained, validated, and tested on 663 normal, 276 polyps, 157 benign, and 146 malignant tumors images. Overall, the model performed at 84.1 ± 4.3% accuracy in the validation set and 80.4 ± 1.7% in the test set. The model correctly identified the presence of a sinonasal mass at 90.5 ± 1.2% accuracy rate. The MLM accuracy performance rates were 86.2 ± 1.0% for polyps and 84.1 ± 1.8% for tumors. Benign and malignant tumor subclassification achieved 87.8 ± 2.1% and 94.0 ± 2.4% accuracy, respectively. Segmentation accuracies for polyps were 72.3% and 72.8% for tumors.
Conclusions: An MLM for nasal endoscopy images can perform with moderate to high accuracy in identifying, classifying, and segmenting sinonasal masses. Performance in future iterations may improve with larger and more diverse training datasets.
Keywords: artificial intelligence; chronic rhinosinusitis; paranasal sinus diseases; polyp.
© 2025 The Author(s). International Forum of Allergy & Rhinology published by Wiley Periodicals LLC on behalf of American Academy of Otolaryngic Allergy and American Rhinologic Society.
Conflict of interest statement
The authors declare they have no conflicts of interest.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

