Topology-based segmentation of 3D confocal images of emerging hematopoietic stem cells in the zebrafish embryo
- PMID: 39776612
- PMCID: PMC11704129
- DOI: 10.1017/S2633903X24000102
Topology-based segmentation of 3D confocal images of emerging hematopoietic stem cells in the zebrafish embryo
Abstract
We develop a novel method for image segmentation of 3D confocal microscopy images of emerging hematopoietic stem cells. The method is based on the theory of persistent homology and uses an optimal threshold to select the most persistent cycles in the persistence diagram. This enables the segmentation of the image's most contrasted and representative shapes. Coupling this segmentation method with a meshing algorithm, we define a pipeline for 3D reconstruction of confocal volumes. Compared to related methods, this approach improves shape segmentation, is more ergonomic to automatize, and has fewer parameters. We apply it to the segmentation of membranes, at subcellular resolution, of cells involved in the endothelial-to-hematopoietic transition (EHT) in the zebrafish embryos.
Keywords: 3D segmentation; cell evolution; meshing; morphogenesis; persistent homology.
© The Author(s) 2024.
Conflict of interest statement
The authors declare no competing interests exist.
Figures
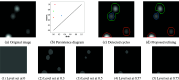







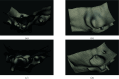
References
-
- Liao P-S, Chen T-S, Chung P-C, et al. (2001) A fast algorithm for multilevel thresholding. Journal of Information Science and Engineering 17(5), 713–727.
-
- Achanta R, Shaji A, Smith K, Lucchi A, Fua P and Süsstrunk S (2012) SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Transactions on Pattern Analysis and Machine Intelligence 34(11), 2274–2282. - PubMed
-
- Chan T and Vese L (1999) An active contour model without edges. In International Conference on Scale-Space Theories in Computer Vision, pp. 141–151. Springer.
-
- Marquez-Neila P, Baumela L and Alvarez L (2013) A morphological approach to curvature-based evolution of curves and surfaces. IEEE Transactions on Pattern Analysis and Machine Intelligence 36(1), 2–17. - PubMed
-
- Schott B, Traub M, Schlagenhauf C, Takamiya M, Antritter T, Bartschat A, Löffler K, Blessing D, Otte JC, Kobitski AY et al. (2018) EmbryoMiner: A new framework for interactive knowledge discovery in large-scale cell tracking data of developing embryos. PLOS Computational Biology 14(4), e1006128. - PMC - PubMed
LinkOut - more resources
Full Text Sources
