The quest for early detection of retinal disease: 3D CycleGAN-based translation of optical coherence tomography into confocal microscopy
- PMID: 39776613
- PMCID: PMC11704141
- DOI: 10.1017/S2633903X24000163
The quest for early detection of retinal disease: 3D CycleGAN-based translation of optical coherence tomography into confocal microscopy
Abstract
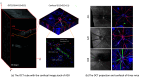
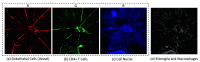
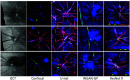
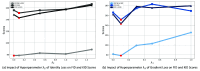
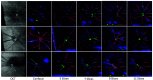
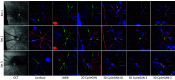
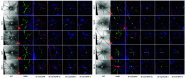
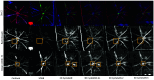
Optical coherence tomography (OCT) and confocal microscopy are pivotal in retinal imaging, offering distinct advantages and limitations. In vivo OCT offers rapid, noninvasive imaging but can suffer from clarity issues and motion artifacts, while ex vivo confocal microscopy, providing high-resolution, cellular-detailed color images, is invasive and raises ethical concerns. To bridge the benefits of both modalities, we propose a novel framework based on unsupervised 3D CycleGAN for translating unpaired in vivo OCT to ex vivo confocal microscopy images. This marks the first attempt to exploit the inherent 3D information of OCT and translate it into the rich, detailed color domain of confocal microscopy. We also introduce a unique dataset, OCT2Confocal, comprising mouse OCT and confocal retinal images, facilitating the development of and establishing a benchmark for cross-modal image translation research. Our model has been evaluated both quantitatively and qualitatively, achieving Fréchet inception distance (FID) scores of 0.766 and Kernel Inception Distance (KID) scores as low as 0.153, and leading subjective mean opinion scores (MOS). Our model demonstrated superior image fidelity and quality with limited data over existing methods. Our approach effectively synthesizes color information from 3D confocal images, closely approximating target outcomes and suggesting enhanced potential for diagnostic and monitoring applications in ophthalmology.
Keywords: CycleGAN; Image-to-image translation; OCT; confocal microscopy; retinal image.
© The Author(s) 2024.
Conflict of interest statement
The authors declare no competing interests exist.
Figures







References
-
- Morano J, Hervella Á, Barreira N, Novo J and Rouco J (2020) Multimodal transfer learning-based approaches for retinal vascular segmentation. arXiv preprint arXiv:2012.10160
LinkOut - more resources
Full Text Sources
