Loss of cped1 does not affect bone and lean tissue in zebrafish
- PMID: 39776615
- PMCID: PMC11701521
- DOI: 10.1093/jbmrpl/ziae159
Loss of cped1 does not affect bone and lean tissue in zebrafish
Abstract
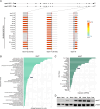
Human genetic studies have nominated cadherin-like and PC-esterase domain-containing 1 (CPED1) as a candidate target gene mediating bone mineral density (BMD) and fracture risk heritability. Recent efforts to define the role of CPED1 in bone in mouse and human models have revealed complex alternative splicing and inconsistent results arising from gene targeting, making its function in bone difficult to interpret. To better understand the role of CPED1 in adult bone mass and morphology, we conducted a comprehensive genetic and phenotypic analysis of cped1 in zebrafish, an emerging model for bone and mineral research. We analyzed two different cped1 mutant lines and performed deep phenotyping to characterize more than 200 measures of adult vertebral, craniofacial, and lean tissue morphology. We also examined alternative splicing of zebrafish cped1 and gene expression in various cell/tissue types. Our studies fail to support an essential role of cped1 in adult zebrafish bone. Specifically, homozygous mutants for both cped1 mutant alleles, which are expected to result in loss-of-function and impact all cped1 isoforms, exhibited no significant differences in the measures examined when compared to their respective wildtype controls, suggesting that cped1 does not significantly contribute to these traits. We identified sequence differences in critical residues of the catalytic triad between the zebrafish and mouse orthologs of CPED1, suggesting that differences in key residues, as well as distinct alternative splicing, could underlie different functions of CPED1 orthologs in the two species. Our studies fail to support a requirement of cped1 in zebrafish bone and lean tissue, adding to evidence that variants at 7q31.31 can act independently of CPED1 to influence BMD and fracture risk.
Keywords: bone; cped1; craniofacial; muscle; zebrafish.
© The Author(s) 2024. Published by Oxford University Press on behalf of the American Society for Bone and Mineral Research.
Conflict of interest statement
None declared.
Figures




Update of
-
Loss of cped1 does not affect bone and lean tissue in zebrafish.bioRxiv [Preprint]. 2024 Aug 17:2024.07.10.601974. doi: 10.1101/2024.07.10.601974. bioRxiv. 2024. Update in: JBMR Plus. 2024 Dec 10;9(2):ziae159. doi: 10.1093/jbmrpl/ziae159. PMID: 39026892 Free PMC article. Updated. Preprint.
References
-
- Cho YS, Go MJ, Kim YJ, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41(5):527–534. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

