Identification of amino acid metabolism‑related genes as diagnostic and prognostic biomarkers in sepsis through machine learning
- PMID: 39776890
- PMCID: PMC11705229
- DOI: 10.3892/etm.2024.12786
Identification of amino acid metabolism‑related genes as diagnostic and prognostic biomarkers in sepsis through machine learning
Abstract
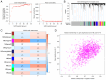
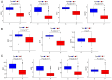
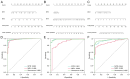
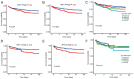
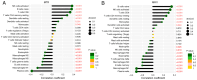
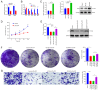
Previous research has highlighted the critical role of amino acid metabolism (AAM) in the pathophysiology of sepsis. The present study aimed to explore the potential diagnostic and prognostic value of AAM-related genes (AAMGs) in sepsis, as well as their underlying molecular mechanisms. Gene expression profiles from the Gene Expression Omnibus (GSE65682, GSE185263 and GSE154918 datasets) were analyzed. Based on weighted gene co-expression network analysis and machine learning algorithms, hub AAMGs were identified in the GSE65682 database. Subsequently, hub AAMGs were evaluated for their expression levels and diagnostic and prognostic significance in sepsis, as well as their interactions with regulatory pathways and role in immune cell infiltration. Additionally, trends in AAMG expression were validated using clinical samples, and their functions in sepsis were confirmed through an in vitro model. In total, four AAMGs were identified, two of which, methionine synthase (MTR) and methionine-R-isomerase 1 (MRI1), demonstrated significant differential expression in the GSE65682, GSE185263 and GSE154918 datasets, which was further validated using clinical samples. A diagnostic nomogram based on MTR and MRI1 expression demonstrated strong diagnostic effectiveness across the three aforementioned databases. Moreover, the expression of both genes were negatively correlated with sepsis prognosis and showed stratified prognostic capabilities. Newly identified pathways included KRAS and IL-2/STAT5 signaling. MTR and MRI1 negatively correlated with the infiltration of inflammatory cells, such as M1 macrophages and neutrophils, and positively correlated with anti-inflammatory cells, such as CD8+ T and dendritic cells. In vitro experiments further demonstrated that overexpression of MTR could mitigate the inhibition of cloning and proliferation induced by LPS and ATP in RAW 264.7 cells. These findings highlighted the potential of MTR and MRI1 as biomarkers for diagnosing and prognosticating sepsis, potentially acting through the regulation of methionine in the pathophysiology of this disease. The present study provided new insights into the role of AAM in the mechanisms underlying sepsis and in the potential development of future targeted therapies.
Keywords: amino acid metabolism; disease markers; inflammation; methionine synthase; methionine-R-isomerase 1; outcome prediction; sepsis.
Copyright: © 2024 Wen et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Fleischmann C, Scherag A, Adhikari NKJ, Hartog CS, Tsaganos T, Schlattmann P, Angus DC, Reinhart K. Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am J Respir Crit Care Med. 2016;193:259–272. doi: 10.1164/rccm.201504-0781OC. International Forum of Acute Care Trialists. - DOI - PubMed
-
- Dai W, Shen J, Yan J, Bott AJ, Maimouni S, Daguplo HQ, Wang Y, Khayati K, Guo JY, Zhang L, et al. Glutamine synthetase limits β-catenin-mutated liver cancer growth by maintaining nitrogen homeostasis and suppressing mTORC1. J Clin Invest. 2022;132(e161408) doi: 10.1172/JCI161408. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
