Meta-analysis of gut microbiota alterations in patients with irritable bowel syndrome
- PMID: 39777150
- PMCID: PMC11703917
- DOI: 10.3389/fmicb.2024.1492349
Meta-analysis of gut microbiota alterations in patients with irritable bowel syndrome
Abstract
Introduction: Irritable bowel syndrome (IBS) is a common chronic disorder of gastrointestinal function with a high prevalence worldwide. Due to its complex pathogenesis and heterogeneity, there is urrently no consensus in IBS research.
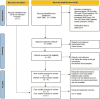
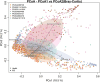
Methods: We collected and uniformly reanalyzed 1167 fecal 16S rRNA gene sequencing samples (623 from IBS patients and 544 from healthy subjects) from 9 studies. Using both a random effects (RE) model and a fixed effects (FE) model, we calculated the odds ratios for metrics including bacterial alpha diversity, beta diversity, common genera and pathways between the IBS and control groups.
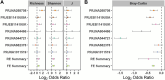
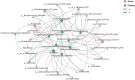
Results: Significantly lower alpha-diversity indexes were observed in IBS patients by random effects model. Twenty-six bacterial genera and twelve predicted pathways were identified with significant odds ratios and classification potentials for IBS patients. Based on these feature, we used transfer learning to enhance the predictive capabilities of our model, which improved model performance by approximately 10%. Moreover, through correlation network analysis, we found that Ruminococcaceae and Christensenellaceae were negatively correlated with vitamin B6 metabolism, which was decreased in the patients with IBS. Ruminococcaceae was also negatively correlated with tyrosine metabolism, which was decreased in the patients with IBS.
Discussion: This study revealed the dysbiosis of fecal bacterial diversity, composition, and predicted pathways of patients with IBS by meta-analysis and identified universal biomarkers for IBS prediction and therapeutic targets.
Keywords: gut microbiota; irritable bowel syndrome; meta-analysis; random forest model; transfer learning.
Copyright © 2024 Li, Li, Xiao, Xu, He, Xiao, Zhang, Cao and Hong.
Conflict of interest statement
HX and JX were employed by company Xiamen Treatgut Biotechnology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Belkova N., Klimenko E., Romanitsa A., Pogodina A., Rychkova L. (2020). Metagenomic 16S rDNA amplicon datasets from adolescents with normal weight, obesity, and obesity with irritable bowel syndrome from eastern Siberia, Russia. Data Brief 32:106141: 106141. doi: 10.1016/j.dib.2020.106141, PMID: - DOI - PMC - PubMed
-
- Carroll I. M., Ringel-Kulka T., Siddle J. P., Ringel Y. (2012). Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol. Motil. 24, 521–30, e248. doi: 10.1111/j.1365-2982.2012.01891.x, PMID: - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

