Urinary TYROBP and HCK as genetic biomarkers for non-invasive diagnosis and therapeutic targeting in IgA nephropathy
- PMID: 39777260
- PMCID: PMC11703869
- DOI: 10.3389/fgene.2024.1516513
Urinary TYROBP and HCK as genetic biomarkers for non-invasive diagnosis and therapeutic targeting in IgA nephropathy
Abstract
Background: IgA nephropathy (IgAN) is a leading cause of renal failure, but its pathogenesis remains unclear, complicating diagnosis and treatment. The invasive nature of renal biopsy highlights the need for non-invasive diagnostic biomarkers. Bulk RNA sequencing (RNA-seq) of urine offers a promising approach for identifying molecular changes relevant to IgAN.
Methods: We performed bulk RNA-seq on 53 urine samples from 11 untreated IgAN patients and 11 healthy controls, integrating these data with public renal RNA-seq, microarray, and scRNA-seq datasets. Machine learning was used to identify key differentially expressed genes, with protein expression validated by immunohistochemistry (IHC) and drug-target interactions explored via molecular docking.
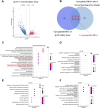
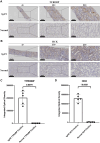
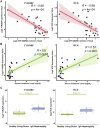
Results: Urine RNA-seq analysis revealed differential expression profiles, from which TYROBP and HCK were identified as key biomarkers using machine learning. These biomarkers were validated in both a test cohort and an external validation cohort, demonstrating strong predictive accuracy. scRNA-seq confirmed their cell-specific expression patterns, correlating with renal function metrics such as GFR and serum creatinine. IHC further validated protein expression, and molecular docking suggested potential therapeutic interactions with IgAN treatments.
Conclusion: TYROBP and HCK are promising non-invasive urinary biomarkers for IgAN. Their predictive accuracy, validated through machine learning, along with IHC confirmation and molecular docking insights, supports their potential for both diagnostic and therapeutic applications in IgAN.
Keywords: HCK; IgA nephropathy; TYROBP; molecular docking; non-invasive biomarkers; urine bulk RNA sequencing.
Copyright © 2024 Xie, Pang, Xie, Tan, Li, Jili, Huang, Zhao, Yuan, Mi, Chen, Ruan, Chen, Li, Hu, Huang, Yang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Barratt J., Lafayette R., Kristensen J., Stone A., Cattran D., Floege J., et al. (2023b). Results from part A of the multi-center, double-blind, randomized, placebo-controlled NefIgArd trial, which evaluated targeted-release formulation of budesonide for the treatment of primary immunoglobulin A nephropathy. Kidney Int. 103 (2), 391–402. 10.1016/j.kint.2022.09.017 - DOI - PubMed
-
- Beckmann M., Wilson T., Lloyd A. J., Torres D., Goios A., Willis N. D., et al. (2020). Challenges associated with the design and deployment of food intake urine biomarker technology for assessment of habitual diet in free-living individuals and populations—a perspective. Front. Nutr. 7, 602515. 10.3389/fnut.2020.602515 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

