Design and in vitro anticancer assessment of a click chemistry-derived dinuclear copper artificial metallo-nuclease
- PMID: 39777469
- PMCID: PMC11705080
- DOI: 10.1093/nar/gkae1250
Design and in vitro anticancer assessment of a click chemistry-derived dinuclear copper artificial metallo-nuclease
Abstract
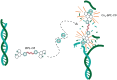
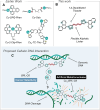
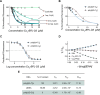
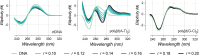
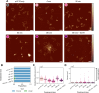
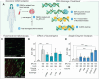
Copper compounds with artificial metallo-nuclease (AMN) activity are mechanistically unique compared to established metallodrugs. Here, we describe the development of a new dinuclear copper AMN, Cu2-BPL-C6 (BPL-C6 = bis-1,10-phenanthroline-carbon-6), prepared using click chemistry that demonstrates site-specific DNA recognition with low micromolar cleavage activity. The BPL-C6 ligand was designed to force two redox-active copper centres-central for enhancing AMN activity-to bind DNA, via two phenanthroline ligands separated by an aliphatic linker. DNA-binding experiments, involving circular dichroism spectroscopy, agarose gel electrophoresis and fluorescence quenching, revealed a preference for binding with adenine-thymine-rich DNA. The oxidative cleavage mechanism of Cu2-BPL-C6 was then elucidated using in vitro molecular and biophysical assays, including in-liquid atomic force microscopy analysis, revealing potent DNA cleavage mediated via superoxide and hydrogen peroxide oxidative pathways. Single-molecule analysis with peripheral blood mononuclear cells identified upregulated single-strand DNA lesions in Cu2-BPL-C6-treated cells. Using specific base excision repair (BER) enzymes, we showed that Endo IV selectively repairs these lesions indicating that the complex generates apurinic and apyrimidinic adducts. Broad spectrum anticancer evaluation of BPL-C6 was performed by the National Cancer Institute's 60 human cell line screen (NCI-60) and revealed selectivity for certain melanoma, breast, colon and non-small cell lung cancer cell lines.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures







References
-
- Watson J.D., Crick F.H.C. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953; 171:737–738. - PubMed
-
- Sigman D.S., Mazumder A., Perrin D.M. Chemical nucleases. Chem. Rev. 1993; 93:2295–2316.
-
- Sigman D.S., Graham D.R., D’Aurora V., Stern A.M. Oxygen-dependent cleavage of DNA by the 1,10-phenanthroline. cuprous complex. Inhibition of Escherichia coli DNA polymerase I. J. Biol. Chem. 1979; 254:12269–12272. - PubMed
-
- Li F., Xie J., Feng F. Copper and zinc complexes of a diaza-crown ether as artificial nucleases for the efficient hydrolytic cleavage of DNA. New J. Chem. 2015; 39:5654–5660.
-
- Zehra S., Tabassum S., Al-Lohedan H.A., Arjmand F. A zwitterionic Zn(II) benzothiazole nanohybrid conjugate as hydrolytic DNA cleavage agent. Inorg. Chem. Commun. 2018; 93:69–72.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

