The role of acetylation and deacetylation in cancer metabolism
- PMID: 39778006
- PMCID: PMC11706801
- DOI: 10.1002/ctm2.70145
The role of acetylation and deacetylation in cancer metabolism
Abstract
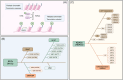
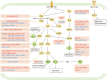
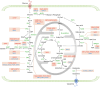
As a hallmark of cancer, metabolic reprogramming adjusts macromolecular synthesis, energy metabolism and redox homeostasis processes to adapt to and promote the complex biological processes of abnormal growth and proliferation. The complexity of metabolic reprogramming lies in its precise regulation by multiple levels and factors, including the interplay of multiple signalling pathways, precise regulation of transcription factors and dynamic adjustments in metabolic enzyme activity. In this complex regulatory network, acetylation and deacetylation, which are important post-translational modifications, regulate key molecules and processes related to metabolic reprogramming by affecting protein function and stability. Dysregulation of acetylation and deacetylation may alter cancer cell metabolic patterns by affecting signalling pathways, transcription factors and metabolic enzyme activity related to metabolic reprogramming, increasing the susceptibility to rapid proliferation and survival. In this review, we focus on discussing how acetylation and deacetylation regulate cancer metabolism, thereby highlighting the central role of these post-translational modifications in metabolic reprogramming, and hoping to provide strong support for the development of novel cancer treatment strategies. KEY POINTS: Protein acetylation and deacetylation are key regulators of metabolic reprogramming in tumour cells. These modifications influence signalling pathways critical for tumour metabolism. They modulate the activity of transcription factors that drive gene expression changes. Metabolic enzymes are also affected, altering cellular metabolism to support tumour growth.
Keywords: acetylation; cancer; deacetylation; metabolic reprogramming.
© 2025 The Author(s). Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646‐674. - PubMed
-
- Icard P, Shulman S, Farhat D, Steyaert JM, Alifano M, Lincet H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells?. Drug Resist Updat. 2018;38:1‐11. - PubMed
-
- Xiao Y, Yu TJ, Xu Y, et al. Emerging therapies in cancer metabolism. Cell Metab. 2023;35(8):1283‐1303. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
